Please be patient as the page loads
|
LOXL4_HUMAN
|
||||||
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LOXL2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994586 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994620 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
LOXL3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.994391 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
LOXL3_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993675 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL4_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL4_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999033 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
DMBT1_HUMAN
|
||||||
| θ value | 1.7756e-72 (rank : 7) | NC score | 0.671503 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | 2.74572e-65 (rank : 8) | NC score | 0.612329 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
NETR_HUMAN
|
||||||
| θ value | 1.01059e-59 (rank : 9) | NC score | 0.414126 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
NETR_MOUSE
|
||||||
| θ value | 5.54529e-58 (rank : 10) | NC score | 0.406020 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
LYOX_MOUSE
|
||||||
| θ value | 7.24236e-58 (rank : 11) | NC score | 0.730445 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
LYOX_HUMAN
|
||||||
| θ value | 1.23536e-57 (rank : 12) | NC score | 0.733322 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
C163B_HUMAN
|
||||||
| θ value | 2.7521e-57 (rank : 13) | NC score | 0.822786 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
C163A_MOUSE
|
||||||
| θ value | 5.19021e-56 (rank : 14) | NC score | 0.824971 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
LOXL1_HUMAN
|
||||||
| θ value | 1.51013e-55 (rank : 15) | NC score | 0.715351 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LOXL1_MOUSE
|
||||||
| θ value | 3.36421e-55 (rank : 16) | NC score | 0.712985 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
C163A_HUMAN
|
||||||
| θ value | 4.39379e-55 (rank : 17) | NC score | 0.824955 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
CD5L_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 18) | NC score | 0.842960 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
CD5L_MOUSE
|
||||||
| θ value | 2.86122e-38 (rank : 19) | NC score | 0.839507 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |
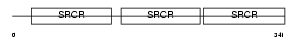
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
CD6_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 20) | NC score | 0.802600 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |
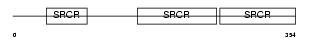
|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
CD6_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 21) | NC score | 0.795742 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |
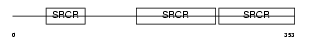
|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
MSRE_MOUSE
|
||||||
| θ value | 2.27234e-27 (rank : 22) | NC score | 0.626502 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
MSRE_HUMAN
|
||||||
| θ value | 2.96777e-27 (rank : 23) | NC score | 0.572987 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
LG3BP_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 24) | NC score | 0.786472 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |
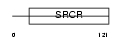
|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
MARCO_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 25) | NC score | 0.489846 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
MARCO_MOUSE
|
||||||
| θ value | 1.2077e-20 (rank : 26) | NC score | 0.390588 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
CFAI_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 27) | NC score | 0.119239 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CFAI_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 28) | NC score | 0.118749 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CD5_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.415764 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |
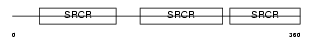
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
K1849_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.021944 (rank : 55) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
TMPS3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.082124 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |
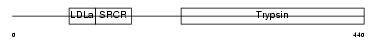
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
TMPS5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.058958 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |
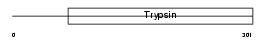
|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
CD5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.389451 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |
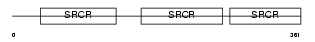
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
TMPS4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.065586 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
ENTK_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.101584 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
TMPS3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.083875 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
TMPS4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.054876 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NRS4, Q5XKQ6, Q6UX37, Q9NZA5 | Gene names | TMPRSS4, TMPRSS3 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Membrane-type serine protease 2) (MT-SP2). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.074314 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TMPS5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.053857 (rank : 45) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H3S3 | Gene names | TMPRSS5 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
ENTK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.086280 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.057399 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
VGFR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | -0.000874 (rank : 57) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35969, O55094, Q61517 | Gene names | Flt1, Emrk2, Flt | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 1 precursor (EC 2.7.10.1) (VEGFR-1) (Tyrosine-protein kinase receptor FLT) (FLT-1) (Embryonic receptor kinase 2). | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.000773 (rank : 56) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
C1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.051657 (rank : 50) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
CORIN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.052946 (rank : 46) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
CS1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.051460 (rank : 52) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
CS1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.052005 (rank : 47) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.084061 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.086800 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
MASP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.051932 (rank : 49) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.051564 (rank : 51) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.051360 (rank : 53) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
ST14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.055559 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
ST14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.057361 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
TMPS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.054798 (rank : 44) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TMPS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.051983 (rank : 48) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TMPS7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.050054 (rank : 54) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
LOXL4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL4_MOUSE
|
||||||
| NC score | 0.999033 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL2_MOUSE
|
||||||
| NC score | 0.994620 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
LOXL2_HUMAN
|
||||||
| NC score | 0.994586 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL3_HUMAN
|
||||||
| NC score | 0.994391 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
LOXL3_MOUSE
|
||||||
| NC score | 0.993675 (rank : 6) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
CD5L_HUMAN
|
||||||
| NC score | 0.842960 (rank : 7) | θ value | 2.34062e-40 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
CD5L_MOUSE
|
||||||
| NC score | 0.839507 (rank : 8) | θ value | 2.86122e-38 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
C163A_MOUSE
|
||||||
| NC score | 0.824971 (rank : 9) | θ value | 5.19021e-56 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163A_HUMAN
|
||||||
| NC score | 0.824955 (rank : 10) | θ value | 4.39379e-55 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163B_HUMAN
|
||||||
| NC score | 0.822786 (rank : 11) | θ value | 2.7521e-57 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
CD6_HUMAN
|
||||||
| NC score | 0.802600 (rank : 12) | θ value | 8.91499e-32 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
CD6_MOUSE
|
||||||
| NC score | 0.795742 (rank : 13) | θ value | 7.80994e-28 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
LG3BP_HUMAN
|
||||||
| NC score | 0.786472 (rank : 14) | θ value | 6.18819e-25 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
LYOX_HUMAN
|
||||||
| NC score | 0.733322 (rank : 15) | θ value | 1.23536e-57 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
LYOX_MOUSE
|
||||||
| NC score | 0.730445 (rank : 16) | θ value | 7.24236e-58 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
LOXL1_HUMAN
|
||||||
| NC score | 0.715351 (rank : 17) | θ value | 1.51013e-55 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LOXL1_MOUSE
|
||||||
| NC score | 0.712985 (rank : 18) | θ value | 3.36421e-55 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
DMBT1_HUMAN
|
||||||
| NC score | 0.671503 (rank : 19) | θ value | 1.7756e-72 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
MSRE_MOUSE
|
||||||
| NC score | 0.626502 (rank : 20) | θ value | 2.27234e-27 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.612329 (rank : 21) | θ value | 2.74572e-65 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
MSRE_HUMAN
|
||||||
| NC score | 0.572987 (rank : 22) | θ value | 2.96777e-27 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
MARCO_HUMAN
|
||||||
| NC score | 0.489846 (rank : 23) | θ value | 3.17815e-21 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
CD5_MOUSE
|
||||||
| NC score | 0.415764 (rank : 24) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
NETR_HUMAN
|
||||||
| NC score | 0.414126 (rank : 25) | θ value | 1.01059e-59 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
NETR_MOUSE
|
||||||
| NC score | 0.406020 (rank : 26) | θ value | 5.54529e-58 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
MARCO_MOUSE
|
||||||
| NC score | 0.390588 (rank : 27) | θ value | 1.2077e-20 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
CD5_HUMAN
|
||||||
| NC score | 0.389451 (rank : 28) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
CFAI_MOUSE
|
||||||
| NC score | 0.119239 (rank : 29) | θ value | 0.00035302 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CFAI_HUMAN
|
||||||
| NC score | 0.118749 (rank : 30) | θ value | 0.00175202 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
ENTK_MOUSE
|
||||||
| NC score | 0.101584 (rank : 31) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.086800 (rank : 32) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
ENTK_HUMAN
|
||||||
| NC score | 0.086280 (rank : 33) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.084061 (rank : 34) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
TMPS3_HUMAN
|
||||||
| NC score | 0.083875 (rank : 35) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
TMPS3_MOUSE
|
||||||
| NC score | 0.082124 (rank : 36) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.074314 (rank : 37) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TMPS4_MOUSE
|
||||||
| NC score | 0.065586 (rank : 38) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
TMPS5_MOUSE
|
||||||
| NC score | 0.058958 (rank : 39) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.057399 (rank : 40) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
ST14_MOUSE
|
||||||
| NC score | 0.057361 (rank : 41) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
ST14_HUMAN
|
||||||
| NC score | 0.055559 (rank : 42) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
TMPS4_HUMAN
|
||||||
| NC score | 0.054876 (rank : 43) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NRS4, Q5XKQ6, Q6UX37, Q9NZA5 | Gene names | TMPRSS4, TMPRSS3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Membrane-type serine protease 2) (MT-SP2). | |||||
|
TMPS2_HUMAN
|
||||||
| NC score | 0.054798 (rank : 44) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TMPS5_HUMAN
|
||||||
| NC score | 0.053857 (rank : 45) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H3S3 | Gene names | TMPRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.052946 (rank : 46) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
CS1B_MOUSE
|
||||||
| NC score | 0.052005 (rank : 47) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
TMPS7_HUMAN
|
||||||
| NC score | 0.051983 (rank : 48) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.051932 (rank : 49) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
C1S_HUMAN
|
||||||
| NC score | 0.051657 (rank : 50) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| NC score | 0.051564 (rank : 51) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
CS1A_MOUSE
|
||||||
| NC score | 0.051460 (rank : 52) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.051360 (rank : 53) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
TMPS7_MOUSE
|
||||||
| NC score | 0.050054 (rank : 54) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
K1849_HUMAN
|
||||||
| NC score | 0.021944 (rank : 55) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.000773 (rank : 56) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
VGFR1_MOUSE
|
||||||
| NC score | -0.000874 (rank : 57) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35969, O55094, Q61517 | Gene names | Flt1, Emrk2, Flt | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 1 precursor (EC 2.7.10.1) (VEGFR-1) (Tyrosine-protein kinase receptor FLT) (FLT-1) (Embryonic receptor kinase 2). | |||||