Please be patient as the page loads
|
CD5L_HUMAN
|
||||||
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |
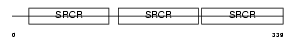
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CD5L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
CD5L_MOUSE
|
||||||
| θ value | 8.33966e-147 (rank : 2) | NC score | 0.995486 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |
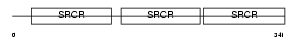
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
DMBT1_HUMAN
|
||||||
| θ value | 3.23591e-74 (rank : 3) | NC score | 0.772569 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
C163A_MOUSE
|
||||||
| θ value | 6.10265e-73 (rank : 4) | NC score | 0.953500 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163B_HUMAN
|
||||||
| θ value | 1.27248e-70 (rank : 5) | NC score | 0.950819 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | 1.83746e-69 (rank : 6) | NC score | 0.702619 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
C163A_HUMAN
|
||||||
| θ value | 2.3998e-69 (rank : 7) | NC score | 0.951535 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
NETR_HUMAN
|
||||||
| θ value | 5.17823e-64 (rank : 8) | NC score | 0.469437 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
NETR_MOUSE
|
||||||
| θ value | 7.47731e-63 (rank : 9) | NC score | 0.456305 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
CD6_HUMAN
|
||||||
| θ value | 6.14528e-49 (rank : 10) | NC score | 0.936369 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |
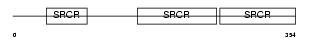
|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
CD6_MOUSE
|
||||||
| θ value | 1.73182e-43 (rank : 11) | NC score | 0.933170 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |
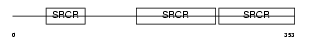
|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
LOXL3_HUMAN
|
||||||
| θ value | 9.50281e-42 (rank : 12) | NC score | 0.840216 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
LOXL2_MOUSE
|
||||||
| θ value | 8.04462e-41 (rank : 13) | NC score | 0.842604 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
LOXL3_MOUSE
|
||||||
| θ value | 2.34062e-40 (rank : 14) | NC score | 0.841043 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL4_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 15) | NC score | 0.842960 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL2_HUMAN
|
||||||
| θ value | 1.51715e-39 (rank : 16) | NC score | 0.840067 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL4_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 17) | NC score | 0.842028 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
MSRE_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 18) | NC score | 0.685331 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
MARCO_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 19) | NC score | 0.535981 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
MSRE_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 20) | NC score | 0.624542 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
MARCO_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 21) | NC score | 0.429375 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
LG3BP_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 22) | NC score | 0.851096 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
CD5_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 23) | NC score | 0.617108 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
CD5_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 24) | NC score | 0.569783 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
CFAI_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 25) | NC score | 0.137234 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CORIN_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 26) | NC score | 0.084231 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
ENTK_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.130321 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
TMPS5_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 28) | NC score | 0.080844 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
ENTK_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.117093 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
CORIN_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.070583 (rank : 52) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
TMPS5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.074777 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H3S3 | Gene names | TMPRSS5 | |||
|
Domain Architecture |
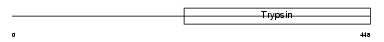
|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
TMPS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.101729 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |
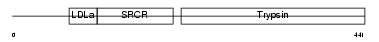
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
TMPS2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.075744 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
CFAI_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.132386 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
TLL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.065989 (rank : 64) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TMPS4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.079623 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |
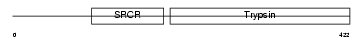
|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
LTB1L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.007019 (rank : 181) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.007031 (rank : 180) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
TLL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.068050 (rank : 58) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TMPS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.098104 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |
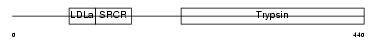
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
XLKD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.010747 (rank : 179) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
FBLN5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.003909 (rank : 182) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVH9, Q541Z7 | Gene names | Fbln5, Dance | |||
|
Domain Architecture |
|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance). | |||||
|
TLL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.066839 (rank : 62) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
UROM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.035621 (rank : 178) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
ZBTB6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | -0.003620 (rank : 183) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||
|
ACRO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.060981 (rank : 77) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |
|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
ACRO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.062125 (rank : 71) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23578 | Gene names | Acr | |||
|
Domain Architecture |
|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
APOA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.052453 (rank : 165) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
BMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.062145 (rank : 70) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.062295 (rank : 69) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
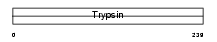
BSSP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.057748 (rank : 108) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZN4, O43342, Q6UXE0 | Gene names | PRSS22, BSSP4, PRSS26 | |||
|
Domain Architecture |
|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
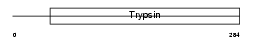
BSSP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.057956 (rank : 106) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ER10 | Gene names | Prss22, Bssp4, Prss26 | |||
|
Domain Architecture |
|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
C1RA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.068173 (rank : 56) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.069367 (rank : 55) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
C1R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.069662 (rank : 54) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
C1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.071139 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
CFAD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.053562 (rank : 150) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00746 | Gene names | CFD, DF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin). | |||||
|
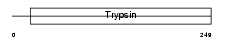
CFAD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.054321 (rank : 141) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P03953, Q61280 | Gene names | Cfd, Adn, Df | |||
|
Domain Architecture |
|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin) (28 kDa adipocyte protein). | |||||
|
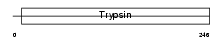
CLCR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.057734 (rank : 110) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99895, O00765, Q9NUH5 | Gene names | CTRC, CLCR | |||
|
Domain Architecture |
|
|||||
| Description | Caldecrin precursor (EC 3.4.21.2) (Chymotrypsin C). | |||||
|
CS1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.071495 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
CS1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.072475 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.051316 (rank : 169) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
CTRB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.056748 (rank : 118) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17538 | Gene names | CTRB1, CTRB | |||
|
Domain Architecture |
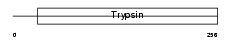
|
|||||
| Description | Chymotrypsinogen B precursor (EC 3.4.21.1) [Contains: Chymotrypsin B chain A; Chymotrypsin B chain B; Chymotrypsin B chain C]. | |||||
|
CTRL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.057047 (rank : 116) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P40313 | Gene names | CTRL, CTRL1 | |||
|
Domain Architecture |
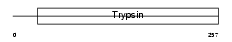
|
|||||
| Description | Chymotrypsin-like protease CTRL-1 precursor (EC 3.4.21.-). | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.116706 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.120423 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
DESC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.059614 (rank : 91) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BZ10, Q8BZ04 | Gene names | Desc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease DESC4 precursor (EC 3.4.21.-) [Contains: Serine protease DESC4 non-catalytic chain; Serine protease DESC4 catalytic chain]. | |||||
|
ELA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.056249 (rank : 123) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNI1, Q5MLF0, Q6DJT0, Q6ISM6 | Gene names | ELA1 | |||
|
Domain Architecture |
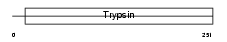
|
|||||
| Description | Elastase-1 precursor (EC 3.4.21.36). | |||||
|
ELA2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.055664 (rank : 128) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08217, Q14243 | Gene names | ELA2A | |||
|
Domain Architecture |
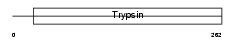
|
|||||
| Description | Elastase-2A precursor (EC 3.4.21.71). | |||||
|
ELA2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.055388 (rank : 133) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05208 | Gene names | Ela2a, Ela-2, Ela2 | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-2A precursor (EC 3.4.21.71) (Elastase-2). | |||||
|
ELA2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.055407 (rank : 132) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08218 | Gene names | ELA2B | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-2B precursor (EC 3.4.21.71). | |||||
|
ELA3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.055829 (rank : 126) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09093, Q9BRW4 | Gene names | ELA3A, ELA3 | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-3A precursor (EC 3.4.21.70) (Elastase IIIA) (Protease E). | |||||
|
ELA3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.055240 (rank : 134) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08861, P11423, Q5VU28, Q5VU29, Q5VU30 | Gene names | ELA3B | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-3B precursor (EC 3.4.21.70) (Elastase IIIB) (Protease E). | |||||
|
ELNE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.050543 (rank : 176) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3UP87, Q61515 | Gene names | Ela2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte elastase precursor (EC 3.4.21.37) (Elastase-2) (Neutrophil elastase). | |||||
|
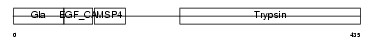
FA10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.054152 (rank : 143) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P00742, Q14340 | Gene names | F10 | |||
|
Domain Architecture |
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) (Stuart- Prower factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
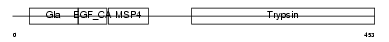
FA10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.053722 (rank : 146) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
FA11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.060088 (rank : 86) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P03951, Q9Y495 | Gene names | F11 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XI precursor (EC 3.4.21.27) (Plasma thromboplastin antecedent) (PTA) (FXI) [Contains: Coagulation factor XIa heavy chain; Coagulation factor XIa light chain]. | |||||
|
FA11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.058864 (rank : 102) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91Y47 | Gene names | F11 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XI precursor (EC 3.4.21.27) (Plasma thromboplastin antecedent) (PTA) (FXI) [Contains: Coagulation factor XIa heavy chain; Coagulation factor XIa light chain]. | |||||
|
FA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.054402 (rank : 140) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
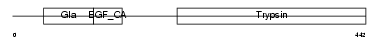
FA7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.052904 (rank : 161) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08709, Q14339, Q5JVF2 | Gene names | F7 | |||
|
Domain Architecture |
|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) (SPCA) (Proconvertin) (Eptacog alfa) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
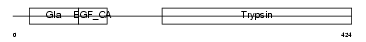
FA7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.053722 (rank : 147) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70375, Q61109 | Gene names | F7, Cf7 | |||
|
Domain Architecture |
|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
GRAK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.051277 (rank : 171) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35205 | Gene names | Gzmk | |||
|
Domain Architecture |
|
|||||
| Description | Granzyme K precursor (EC 3.4.21.-). | |||||
|
HABP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.053416 (rank : 152) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14520, O00663 | Gene names | HABP2, HGFAL, PHBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) (Hepatocyte growth factor activator-like protein) (Factor VII-activating protease) (Factor seven-activating protease) (FSAP) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
HABP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.053048 (rank : 158) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0D2 | Gene names | Habp2, Phbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
HEPS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.061230 (rank : 75) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05981 | Gene names | HPN, TMPRSS1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease hepsin (EC 3.4.21.-) (Transmembrane protease, serine 1) [Contains: Serine protease hepsin non-catalytic chain; Serine protease hepsin catalytic chain]. | |||||
|
HEPS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.060710 (rank : 78) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35453, Q9CW97 | Gene names | Hpn | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease hepsin (EC 3.4.21.-) [Contains: Serine protease hepsin non-catalytic chain; Serine protease hepsin catalytic chain]. | |||||
|
HGFA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.054897 (rank : 137) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
HGFA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053586 (rank : 149) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
HPTR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051294 (rank : 170) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00739, Q7LE20, Q92658, Q92659, Q9ULB0 | Gene names | HPR | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin-related protein precursor. | |||||
|
HPT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.051409 (rank : 168) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00738, P00737, Q2PP15, Q3B7J0, Q6LBY9 | Gene names | HP | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
HPT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.051242 (rank : 172) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61646 | Gene names | Hp | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
KLK10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.053703 (rank : 148) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43240, Q99920, Q9GZW9 | Gene names | KLK10, NES1, PRRSL1 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-10 precursor (EC 3.4.21.-) (Protease serine-like 1) (Normal epithelial cell-specific 1). | |||||
|
KLK12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.050020 (rank : 177) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKR0, Q9UKR1 | Gene names | KLK12, KLKL5 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-12 precursor (EC 3.4.21.-) (Kallikrein-like protein 5) (KLK-L5). | |||||
|
KLK13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.053378 (rank : 154) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKR3, Q9Y433 | Gene names | KLK13, KLKL4 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-13 precursor (EC 3.4.21.-) (Kallikrein-like protein 4) (KLK-L4). | |||||
|
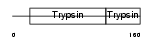
KLK15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.050814 (rank : 174) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H2R5, Q15358, Q9H2R3, Q9H2R4, Q9H2R6, Q9HBG9 | Gene names | KLK15 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-15 precursor (EC 3.4.21.-) (ACO protease). | |||||
|
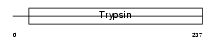
KLK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.053386 (rank : 153) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92876 | Gene names | KLK6, PRSS18, PRSS9 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-6 precursor (EC 3.4.21.-) (Protease M) (Neurosin) (Zyme) (SP59). | |||||
|
KLK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.050642 (rank : 175) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91VE3, Q9R048 | Gene names | Klk7, Prss6, Scce | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kallikrein-7 precursor (EC 3.4.21.-) (Stratum corneum chymotryptic enzyme) (Thymopsin). | |||||
|
KLKB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.057873 (rank : 107) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P03952 | Gene names | KLKB1, KLK3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
KLKB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.059126 (rank : 98) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26262 | Gene names | Klkb1, Klk3, Pk | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
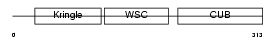
KREM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.055186 (rank : 135) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MU8, Q5TIB9, Q6P3X6, Q9BY70, Q9UGS5, Q9UGU1 | Gene names | KREMEN1, KREMEN, KRM1 | |||
|
Domain Architecture |
|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
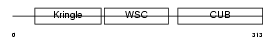
KREM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.057732 (rank : 111) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |
|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
LOXL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.287334 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LOXL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.286341 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
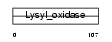
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
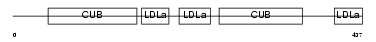
LRP12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.061332 (rank : 74) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
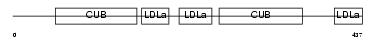
LRP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.061409 (rank : 73) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
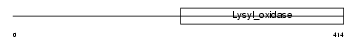
LYOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.302025 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
LYOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.299729 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |
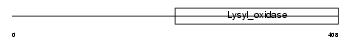
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
MASP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.073044 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.072574 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.071373 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MASP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.068031 (rank : 59) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MCPT6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.056156 (rank : 124) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P21845, Q61962 | Gene names | Mcpt6 | |||
|
Domain Architecture |
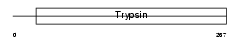
|
|||||
| Description | Mast cell protease 6 precursor (EC 3.4.21.59) (MMCP-6) (Tryptase). | |||||
|
MFRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.061752 (rank : 72) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.062401 (rank : 68) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NETO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.058017 (rank : 105) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.057308 (rank : 115) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.059128 (rank : 97) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.059454 (rank : 94) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NRPN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051561 (rank : 167) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60259, Q9HCB3, Q9UIL9, Q9UQ47 | Gene names | KLK8, NRPN, PRSS19, TADG14 | |||
|
Domain Architecture |
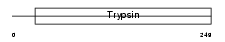
|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8) (Ovasin) (Serine protease TADG-14) (Tumor-associated differentially expressed gene 14 protein). | |||||
|
NRPN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.052563 (rank : 163) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61955 | Gene names | Klk8, Nrpn, Prss19 | |||
|
Domain Architecture |
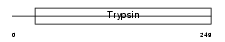
|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.060104 (rank : 85) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.061072 (rank : 76) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.066261 (rank : 63) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.067081 (rank : 61) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PLMN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.053101 (rank : 157) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00747, Q15146, Q6PA00 | Gene names | PLG | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
PLMN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052940 (rank : 159) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20918, Q8CIS2, Q91WJ5 | Gene names | Plg | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
POLS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.056547 (rank : 120) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5K4E3, Q8NBY4 | Gene names | PRSS36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
POLS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.054944 (rank : 136) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5K2P8 | Gene names | Prss36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
PROC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.054876 (rank : 138) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P04070, Q15189, Q15190, Q16001 | Gene names | PROC | |||
|
Domain Architecture |
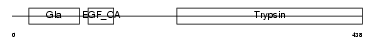
|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
PROC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.053376 (rank : 155) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |
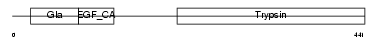
|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
PRS27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.057436 (rank : 114) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BQR3 | Gene names | PRSS27, MPN | |||
|
Domain Architecture |
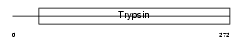
|
|||||
| Description | Protease serine 27 precursor (EC 3.4.21.-) (Marapsin) (Pancreasin) (Channel-activating protease 2) (CAPH2). | |||||
|
PRS27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.057626 (rank : 112) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJR6 | Gene names | Prss27, Mpn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protease serine 27 precursor (EC 3.4.21.-) (Marapsin) (Pancreasin). | |||||
|
PRSS8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.055706 (rank : 127) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16651, Q9UCA3 | Gene names | PRSS8 | |||
|
Domain Architecture |
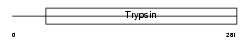
|
|||||
| Description | Prostasin precursor (EC 3.4.21.-) [Contains: Prostasin light chain; Prostasin heavy chain]. | |||||
|
PRSS8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.056619 (rank : 119) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ESD1 | Gene names | Prss8, Cap1 | |||
|
Domain Architecture |
|
|||||
| Description | Prostasin precursor (EC 3.4.21.-) (Channel-activating protease 1) [Contains: Prostasin light chain; Prostasin heavy chain]. | |||||
|
ST14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.077269 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
ST14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.079467 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
TECTB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.056518 (rank : 122) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PL2 | Gene names | TECTB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
TECTB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.056545 (rank : 121) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08524 | Gene names | Tectb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
TEST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.060607 (rank : 82) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6M0, Q9NS34, Q9P2V6 | Gene names | PRSS21, ESP1, TEST1 | |||
|
Domain Architecture |
|
|||||
| Description | Testisin precursor (EC 3.4.21.-) (Eosinophil serine protease 1) (ESP- 1). | |||||
|
TEST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.057476 (rank : 113) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JHJ7, Q9DA14 | Gene names | Prss21 | |||
|
Domain Architecture |
|
|||||
| Description | Testisin precursor (EC 3.4.21.-) (Tryptase 4). | |||||
|
TGBR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.056111 (rank : 125) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03167, Q5T2T4, Q5U731, Q9UGI2 | Gene names | TGFBR3 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
TGBR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.054706 (rank : 139) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88393, Q6NS72 | Gene names | Tgfbr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
THRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.053527 (rank : 151) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00734 | Gene names | F2 | |||
|
Domain Architecture |

|
|||||
| Description | Prothrombin precursor (EC 3.4.21.5) (Coagulation factor II) [Contains: Activation peptide fragment 1; Activation peptide fragment 2; Thrombin light chain; Thrombin heavy chain]. | |||||
|
THRB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.052769 (rank : 162) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19221 | Gene names | F2, Cf2 | |||
|
Domain Architecture |

|
|||||
| Description | Prothrombin precursor (EC 3.4.21.5) (Coagulation factor II) [Contains: Activation peptide fragment 1; Activation peptide fragment 2; Thrombin light chain; Thrombin heavy chain]. | |||||
|
TLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.064153 (rank : 67) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TM11D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.059566 (rank : 92) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60235 | Gene names | TMPRSS11D, HAT | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 11D precursor (EC 3.4.21.-) (Airway trypsin-like protease) [Contains: Transmembrane protease, serine 11D non-catalytic chain; Transmembrane protease, serine 11D catalytic chain]. | |||||
|
TM11D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.060062 (rank : 87) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHK8, Q7TNX3, Q8VDV1 | Gene names | Tmprss11d, Mat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11D precursor (EC 3.4.21.-) (Airway trypsin-like protease) (AT) (Adrenal secretory serine protease) (AsP) [Contains: Transmembrane protease, serine 11D non-catalytic chain; Transmembrane protease, serine 11D catalytic chain]. | |||||
|
TM11E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.060393 (rank : 84) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UL52, Q6UW31 | Gene names | TMPRSS11E, DESC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11E precursor (EC 3.4.21.-) (Serine protease DESC1) [Contains: Transmembrane protease, serine 11E non- catalytic chain; Transmembrane protease, serine 11E catalytic chain]. | |||||
|
TM11E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.059040 (rank : 99) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5S248, Q8BM10 | Gene names | Tmprss11e, Desc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11E precursor (EC 3.4.21.-) (Serine protease DESC1) [Contains: Transmembrane protease, serine 11E non- catalytic chain; Transmembrane protease, serine 11E catalytic chain]. | |||||
|
TMPS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.065762 (rank : 65) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TMPS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.067399 (rank : 60) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NRS4, Q5XKQ6, Q6UX37, Q9NZA5 | Gene names | TMPRSS4, TMPRSS3 | |||
|
Domain Architecture |
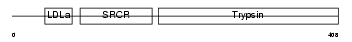
|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Membrane-type serine protease 2) (MT-SP2). | |||||
|
TMPS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.059919 (rank : 89) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
TMPS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.059347 (rank : 96) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
TMPS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.070577 (rank : 53) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TMPS7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.068053 (rank : 57) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TMPS9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.058930 (rank : 101) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z410, Q6ZND6, Q7Z411 | Gene names | TMPRSS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserase-I) (Polyserine protease 1) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
TMPS9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.058561 (rank : 103) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P69525 | Gene names | Tmprss9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserine protease 1) (Polyserase-I) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.073205 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.091757 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TMSP8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.059958 (rank : 88) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYZ9, Q91XC4 | Gene names | Tmprss8, Disp | |||
|
Domain Architecture |
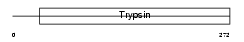
|
|||||
| Description | Transmembrane serine protease 8 precursor (EC 3.4.21.-) (Distal intestinal serine protease). | |||||
|
TPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.059481 (rank : 93) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P00750, Q15103, Q7Z7N2, Q86YK8, Q9BU99 | Gene names | PLAT | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) (Alteplase) (Reteplase) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||
|
TPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.060688 (rank : 80) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11214, Q91VP2 | Gene names | Plat | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||
|
TRY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.052558 (rank : 164) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07477, Q92955, Q9HAN4, Q9HAN5, Q9HAN6, Q9HAN7 | Gene names | PRSS1, TRP1, TRY1, TRYP1 | |||
|
Domain Architecture |
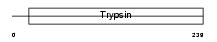
|
|||||
| Description | Trypsin-1 precursor (EC 3.4.21.4) (Trypsin I) (Cationic trypsinogen). | |||||
|
TRY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.052297 (rank : 166) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07478 | Gene names | PRSS2, TRY2, TRYP2 | |||
|
Domain Architecture |
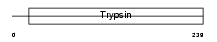
|
|||||
| Description | Trypsin-2 precursor (EC 3.4.21.4) (Trypsin II) (Anionic trypsinogen). | |||||
|
TRY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.052908 (rank : 160) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07146 | Gene names | Prss2, Try2 | |||
|
Domain Architecture |
|
|||||
| Description | Anionic trypsin-2 precursor (EC 3.4.21.4) (Anionic trypsin II) (Pretrypsinogen II). | |||||
|
TRY3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.053194 (rank : 156) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35030, P15951, Q15665, Q9UQV3 | Gene names | PRSS3, PRSS4, TRY3, TRY4 | |||
|
Domain Architecture |
|
|||||
| Description | Trypsin-3 precursor (EC 3.4.21.4) (Trypsin III) (Brain trypsinogen) (Mesotrypsinogen) (Trypsin IV). | |||||
|
TRYA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.055606 (rank : 129) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15157, Q9H2Y5, Q9UQI1 | Gene names | TPSAB1, TPS1 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase alpha-1 precursor (EC 3.4.21.59) (Tryptase-1). | |||||
|
TRYB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.055496 (rank : 130) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15661, Q15663, Q9H2Y4 | Gene names | TPSAB1, TPSB1 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase beta-1 precursor (EC 3.4.21.59) (Tryptase-1) (Tryptase I). | |||||
|
TRYB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.054200 (rank : 142) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02844 | Gene names | Tpsab1, Mcpt7 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase precursor (EC 3.4.21.59) (Mast cell protease 7) (MMCP-7) (Tryptase alpha/beta-1). | |||||
|
TRYB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.055496 (rank : 131) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20231, O95827, Q15664, Q9UQI6, Q9UQI7 | Gene names | TPSB2, TPS2 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase beta-2 precursor (EC 3.4.21.59) (Tryptase-2) (Tryptase II). | |||||
|
TRYD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.053994 (rank : 145) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZJ3, O95824, Q8TDI6, Q96L36, Q96RZ5, Q9H2Y6, Q9UQI8 | Gene names | TPSD1 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase delta precursor (EC 3.4.21.59) (Delta tryptase) (Mast cell mMCP-7-like) (Tryptase-3) (HmMCP-3-like tryptase III). | |||||
|
TRYG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.058189 (rank : 104) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRR2, Q96RZ8, Q9C015, Q9NRQ8, Q9UBB2 | Gene names | TPSG1, TMT | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase gamma precursor (EC 3.4.21.-) (Transmembrane tryptase) [Contains: Tryptase gamma light chain; Tryptase gamma heavy chain]. | |||||
|
TRYG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.059007 (rank : 100) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QUL7 | Gene names | Tpsg1, Tmt | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase gamma precursor (EC 3.4.21.-) (Transmembrane tryptase) [Contains: Tryptase gamma light chain; Tryptase gamma heavy chain]. | |||||
|
TSG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.060704 (rank : 79) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.060605 (rank : 83) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
TSP50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.060670 (rank : 81) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UI38 | Gene names | TSP50 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protease-like protein 50 precursor. | |||||
|
UROK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.059698 (rank : 90) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P00749, Q15844, Q16618, Q969W6 | Gene names | PLAU | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase-type plasminogen activator precursor (EC 3.4.21.73) (uPA) (U-plasminogen activator) [Contains: Urokinase-type plasminogen activator long chain A; Urokinase-type plasminogen activator short chain A; Urokinase-type plasminogen activator chain B]. | |||||
|
UROK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.056986 (rank : 117) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06869 | Gene names | Plau | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase-type plasminogen activator precursor (EC 3.4.21.73) (uPA) (U-plasminogen activator) [Contains: Urokinase-type plasminogen activator long chain A; Urokinase-type plasminogen activator short chain A; Urokinase-type plasminogen activator chain B]. | |||||
|
ZP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.065102 (rank : 66) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
ZP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.059358 (rank : 95) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20239 | Gene names | Zp2, Zp-2, Zpa | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
ZP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.050938 (rank : 173) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21754, Q06633 | Gene names | ZP3, ZP3A, ZPC | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Zona pellucida protein C) (Sperm receptor) (ZP3A/ZP3B). | |||||
|
ZP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.054065 (rank : 144) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10761 | Gene names | Zp3, Zp-3, Zpc | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Sperm receptor) (Zona pellucida protein C). | |||||
|
ZP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.057747 (rank : 109) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12836 | Gene names | ZP4, ZPB | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 4 precursor (Zona pellucida sperm-binding protein B). | |||||
|
CD5L_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
CD5L_MOUSE
|
||||||
| NC score | 0.995486 (rank : 2) | θ value | 8.33966e-147 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
C163A_MOUSE
|
||||||
| NC score | 0.953500 (rank : 3) | θ value | 6.10265e-73 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163A_HUMAN
|
||||||
| NC score | 0.951535 (rank : 4) | θ value | 2.3998e-69 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163B_HUMAN
|
||||||
| NC score | 0.950819 (rank : 5) | θ value | 1.27248e-70 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
CD6_HUMAN
|
||||||
| NC score | 0.936369 (rank : 6) | θ value | 6.14528e-49 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
CD6_MOUSE
|
||||||
| NC score | 0.933170 (rank : 7) | θ value | 1.73182e-43 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
LG3BP_HUMAN
|
||||||
| NC score | 0.851096 (rank : 8) | θ value | 2.35696e-16 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
LOXL4_HUMAN
|
||||||
| NC score | 0.842960 (rank : 9) | θ value | 2.34062e-40 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL2_MOUSE
|
||||||
| NC score | 0.842604 (rank : 10) | θ value | 8.04462e-41 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
LOXL4_MOUSE
|
||||||
| NC score | 0.842028 (rank : 11) | θ value | 4.88048e-38 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL3_MOUSE
|
||||||
| NC score | 0.841043 (rank : 12) | θ value | 2.34062e-40 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL3_HUMAN
|
||||||
| NC score | 0.840216 (rank : 13) | θ value | 9.50281e-42 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
LOXL2_HUMAN
|
||||||
| NC score | 0.840067 (rank : 14) | θ value | 1.51715e-39 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
DMBT1_HUMAN
|
||||||
| NC score | 0.772569 (rank : 15) | θ value | 3.23591e-74 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.702619 (rank : 16) | θ value | 1.83746e-69 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
MSRE_MOUSE
|
||||||
| NC score | 0.685331 (rank : 17) | θ value | 1.42661e-21 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
MSRE_HUMAN
|
||||||
| NC score | 0.624542 (rank : 18) | θ value | 1.5773e-20 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
CD5_MOUSE
|
||||||
| NC score | 0.617108 (rank : 19) | θ value | 1.58096e-12 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
CD5_HUMAN
|
||||||
| NC score | 0.569783 (rank : 20) | θ value | 2.61198e-07 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
MARCO_HUMAN
|
||||||
| NC score | 0.535981 (rank : 21) | θ value | 5.42112e-21 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
NETR_HUMAN
|
||||||
| NC score | 0.469437 (rank : 22) | θ value | 5.17823e-64 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
NETR_MOUSE
|
||||||
| NC score | 0.456305 (rank : 23) | θ value | 7.47731e-63 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
MARCO_MOUSE
|
||||||
| NC score | 0.429375 (rank : 24) | θ value | 4.58923e-20 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
LYOX_HUMAN
|
||||||
| NC score | 0.302025 (rank : 25) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
LYOX_MOUSE
|
||||||
| NC score | 0.299729 (rank : 26) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
LOXL1_HUMAN
|
||||||
| NC score | 0.287334 (rank : 27) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LOXL1_MOUSE
|
||||||
| NC score | 0.286341 (rank : 28) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
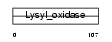
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
CFAI_HUMAN
|
||||||
| NC score | 0.137234 (rank : 29) | θ value | 0.000461057 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CFAI_MOUSE
|
||||||
| NC score | 0.132386 (rank : 30) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
ENTK_MOUSE
|
||||||
| NC score | 0.130321 (rank : 31) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.120423 (rank : 32) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
ENTK_HUMAN
|
||||||
| NC score | 0.117093 (rank : 33) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.116706 (rank : 34) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
TMPS3_HUMAN
|
||||||
| NC score | 0.101729 (rank : 35) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
TMPS3_MOUSE
|
||||||
| NC score | 0.098104 (rank : 36) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.091757 (rank : 37) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.084231 (rank : 38) | θ value | 0.00390308 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
TMPS5_MOUSE
|
||||||
| NC score | 0.080844 (rank : 39) | θ value | 0.0193708 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
TMPS4_MOUSE
|
||||||
| NC score | 0.079623 (rank : 40) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
ST14_MOUSE
|
||||||
| NC score | 0.079467 (rank : 41) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
ST14_HUMAN
|
||||||
| NC score | 0.077269 (rank : 42) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
TMPS2_HUMAN
|
||||||
| NC score | 0.075744 (rank : 43) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TMPS5_HUMAN
|
||||||
| NC score | 0.074777 (rank : 44) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H3S3 | Gene names | TMPRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.073205 (rank : 45) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.073044 (rank : 46) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| NC score | 0.072574 (rank : 47) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
CS1B_MOUSE
|
||||||
| NC score | 0.072475 (rank : 48) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
CS1A_MOUSE
|
||||||
| NC score | 0.071495 (rank : 49) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.071373 (rank : 50) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
C1S_HUMAN
|
||||||
| NC score | 0.071139 (rank : 51) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
CORIN_HUMAN
|
||||||
| NC score | 0.070583 (rank : 52) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
TMPS7_HUMAN
|
||||||
| NC score | 0.070577 (rank : 53) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
C1R_HUMAN
|
||||||
| NC score | 0.069662 (rank : 54) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| NC score | 0.069367 (rank : 55) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
C1RA_MOUSE
|
||||||
| NC score | 0.068173 (rank : 56) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
TMPS7_MOUSE
|
||||||
| NC score | 0.068053 (rank : 57) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TLL1_MOUSE
|
||||||
| NC score | 0.068050 (rank : 58) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
MASP2_MOUSE
|
||||||
| NC score | 0.068031 (rank : 59) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
TMPS4_HUMAN
|
||||||
| NC score | 0.067399 (rank : 60) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NRS4, Q5XKQ6, Q6UX37, Q9NZA5 | Gene names | TMPRSS4, TMPRSS3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Membrane-type serine protease 2) (MT-SP2). | |||||
|
PCOC2_MOUSE
|
||||||
| NC score | 0.067081 (rank : 61) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
TLL1_HUMAN
|
||||||
| NC score | 0.066839 (rank : 62) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
PCOC2_HUMAN
|
||||||
| NC score | 0.066261 (rank : 63) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
TLL2_MOUSE
|
||||||
| NC score | 0.065989 (rank : 64) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TMPS2_MOUSE
|
||||||
| NC score | 0.065762 (rank : 65) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
ZP2_HUMAN
|
||||||
| NC score | 0.065102 (rank : 66) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
TLL2_HUMAN
|
||||||
| NC score | 0.064153 (rank : 67) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.062401 (rank : 68) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
BMP1_MOUSE
|
||||||
| NC score | 0.062295 (rank : 69) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_HUMAN
|
||||||
| NC score | 0.062145 (rank : 70) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
ACRO_MOUSE
|
||||||
| NC score | 0.062125 (rank : 71) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23578 | Gene names | Acr | |||
|
Domain Architecture |

|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.061752 (rank : 72) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.061409 (rank : 73) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LRP12_HUMAN
|
||||||
| NC score | 0.061332 (rank : 74) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
HEPS_HUMAN
|
||||||
| NC score | 0.061230 (rank : 75) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05981 | Gene names | HPN, TMPRSS1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease hepsin (EC 3.4.21.-) (Transmembrane protease, serine 1) [Contains: Serine protease hepsin non-catalytic chain; Serine protease hepsin catalytic chain]. | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.061072 (rank : 76) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
ACRO_HUMAN
|
||||||
| NC score | 0.060981 (rank : 77) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |

|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
HEPS_MOUSE
|
||||||
| NC score | 0.060710 (rank : 78) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35453, Q9CW97 | Gene names | Hpn | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease hepsin (EC 3.4.21.-) [Contains: Serine protease hepsin non-catalytic chain; Serine protease hepsin catalytic chain]. | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.060704 (rank : 79) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TPA_MOUSE
|
||||||
| NC score | 0.060688 (rank : 80) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11214, Q91VP2 | Gene names | Plat | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||
|
TSP50_HUMAN
|
||||||
| NC score | 0.060670 (rank : 81) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UI38 | Gene names | TSP50 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protease-like protein 50 precursor. | |||||
|
TEST_HUMAN
|
||||||
| NC score | 0.060607 (rank : 82) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6M0, Q9NS34, Q9P2V6 | Gene names | PRSS21, ESP1, TEST1 | |||
|
Domain Architecture |

|
|||||
| Description | Testisin precursor (EC 3.4.21.-) (Eosinophil serine protease 1) (ESP- 1). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.060605 (rank : 83) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
TM11E_HUMAN
|
||||||
| NC score | 0.060393 (rank : 84) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UL52, Q6UW31 | Gene names | TMPRSS11E, DESC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11E precursor (EC 3.4.21.-) (Serine protease DESC1) [Contains: Transmembrane protease, serine 11E non- catalytic chain; Transmembrane protease, serine 11E catalytic chain]. | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.060104 (rank : 85) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
FA11_HUMAN
|
||||||
| NC score | 0.060088 (rank : 86) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P03951, Q9Y495 | Gene names | F11 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XI precursor (EC 3.4.21.27) (Plasma thromboplastin antecedent) (PTA) (FXI) [Contains: Coagulation factor XIa heavy chain; Coagulation factor XIa light chain]. | |||||
|
TM11D_MOUSE
|
||||||
| NC score | 0.060062 (rank : 87) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHK8, Q7TNX3, Q8VDV1 | Gene names | Tmprss11d, Mat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11D precursor (EC 3.4.21.-) (Airway trypsin-like protease) (AT) (Adrenal secretory serine protease) (AsP) [Contains: Transmembrane protease, serine 11D non-catalytic chain; Transmembrane protease, serine 11D catalytic chain]. | |||||
|
TMSP8_MOUSE
|
||||||
| NC score | 0.059958 (rank : 88) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYZ9, Q91XC4 | Gene names | Tmprss8, Disp | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane serine protease 8 precursor (EC 3.4.21.-) (Distal intestinal serine protease). | |||||
|
TMPS6_HUMAN
|
||||||
| NC score | 0.059919 (rank : 89) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
UROK_HUMAN
|
||||||
| NC score | 0.059698 (rank : 90) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P00749, Q15844, Q16618, Q969W6 | Gene names | PLAU | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase-type plasminogen activator precursor (EC 3.4.21.73) (uPA) (U-plasminogen activator) [Contains: Urokinase-type plasminogen activator long chain A; Urokinase-type plasminogen activator short chain A; Urokinase-type plasminogen activator chain B]. | |||||
|
DESC4_MOUSE
|
||||||
| NC score | 0.059614 (rank : 91) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BZ10, Q8BZ04 | Gene names | Desc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease DESC4 precursor (EC 3.4.21.-) [Contains: Serine protease DESC4 non-catalytic chain; Serine protease DESC4 catalytic chain]. | |||||
|
TM11D_HUMAN
|
||||||
| NC score | 0.059566 (rank : 92) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60235 | Gene names | TMPRSS11D, HAT | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 11D precursor (EC 3.4.21.-) (Airway trypsin-like protease) [Contains: Transmembrane protease, serine 11D non-catalytic chain; Transmembrane protease, serine 11D catalytic chain]. | |||||
|
TPA_HUMAN
|
||||||
| NC score | 0.059481 (rank : 93) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P00750, Q15103, Q7Z7N2, Q86YK8, Q9BU99 | Gene names | PLAT | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) (Alteplase) (Reteplase) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||
|
NETO2_MOUSE
|
||||||
| NC score | 0.059454 (rank : 94) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
ZP2_MOUSE
|
||||||
| NC score | 0.059358 (rank : 95) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20239 | Gene names | Zp2, Zp-2, Zpa | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
TMPS6_MOUSE
|
||||||
| NC score | 0.059347 (rank : 96) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
NETO2_HUMAN
|
||||||
| NC score | 0.059128 (rank : 97) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
KLKB1_MOUSE
|
||||||
| NC score | 0.059126 (rank : 98) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26262 | Gene names | Klkb1, Klk3, Pk | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
TM11E_MOUSE
|
||||||
| NC score | 0.059040 (rank : 99) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5S248, Q8BM10 | Gene names | Tmprss11e, Desc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11E precursor (EC 3.4.21.-) (Serine protease DESC1) [Contains: Transmembrane protease, serine 11E non- catalytic chain; Transmembrane protease, serine 11E catalytic chain]. | |||||
|
TRYG1_MOUSE
|
||||||
| NC score | 0.059007 (rank : 100) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QUL7 | Gene names | Tpsg1, Tmt | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase gamma precursor (EC 3.4.21.-) (Transmembrane tryptase) [Contains: Tryptase gamma light chain; Tryptase gamma heavy chain]. | |||||
|
TMPS9_HUMAN
|
||||||
| NC score | 0.058930 (rank : 101) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z410, Q6ZND6, Q7Z411 | Gene names | TMPRSS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserase-I) (Polyserine protease 1) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
FA11_MOUSE
|
||||||
| NC score | 0.058864 (rank : 102) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91Y47 | Gene names | F11 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XI precursor (EC 3.4.21.27) (Plasma thromboplastin antecedent) (PTA) (FXI) [Contains: Coagulation factor XIa heavy chain; Coagulation factor XIa light chain]. | |||||
|
TMPS9_MOUSE
|
||||||
| NC score | 0.058561 (rank : 103) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P69525 | Gene names | Tmprss9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserine protease 1) (Polyserase-I) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
TRYG1_HUMAN
|
||||||
| NC score | 0.058189 (rank : 104) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRR2, Q96RZ8, Q9C015, Q9NRQ8, Q9UBB2 | Gene names | TPSG1, TMT | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase gamma precursor (EC 3.4.21.-) (Transmembrane tryptase) [Contains: Tryptase gamma light chain; Tryptase gamma heavy chain]. | |||||
|
NETO1_HUMAN
|
||||||
| NC score | 0.058017 (rank : 105) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
BSSP4_MOUSE
|
||||||
| NC score | 0.057956 (rank : 106) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ER10 | Gene names | Prss22, Bssp4, Prss26 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
KLKB1_HUMAN
|
||||||
| NC score | 0.057873 (rank : 107) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P03952 | Gene names | KLKB1, KLK3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
BSSP4_HUMAN
|
||||||
| NC score | 0.057748 (rank : 108) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZN4, O43342, Q6UXE0 | Gene names | PRSS22, BSSP4, PRSS26 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
ZP4_HUMAN
|
||||||
| NC score | 0.057747 (rank : 109) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12836 | Gene names | ZP4, ZPB | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 4 precursor (Zona pellucida sperm-binding protein B). | |||||
|
CLCR_HUMAN
|
||||||
| NC score | 0.057734 (rank : 110) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99895, O00765, Q9NUH5 | Gene names | CTRC, CLCR | |||
|
Domain Architecture |

|
|||||
| Description | Caldecrin precursor (EC 3.4.21.2) (Chymotrypsin C). | |||||
|
KREM1_MOUSE
|
||||||
| NC score | 0.057732 (rank : 111) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
PRS27_MOUSE
|
||||||
| NC score | 0.057626 (rank : 112) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJR6 | Gene names | Prss27, Mpn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protease serine 27 precursor (EC 3.4.21.-) (Marapsin) (Pancreasin). | |||||
|
TEST_MOUSE
|
||||||
| NC score | 0.057476 (rank : 113) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JHJ7, Q9DA14 | Gene names | Prss21 | |||
|
Domain Architecture |

|
|||||
| Description | Testisin precursor (EC 3.4.21.-) (Tryptase 4). | |||||
|
PRS27_HUMAN
|
||||||
| NC score | 0.057436 (rank : 114) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BQR3 | Gene names | PRSS27, MPN | |||
|
Domain Architecture |

|
|||||
| Description | Protease serine 27 precursor (EC 3.4.21.-) (Marapsin) (Pancreasin) (Channel-activating protease 2) (CAPH2). | |||||
|
NETO1_MOUSE
|
||||||
| NC score | 0.057308 (rank : 115) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
CTRL_HUMAN
|
||||||
| NC score | 0.057047 (rank : 116) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P40313 | Gene names | CTRL, CTRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chymotrypsin-like protease CTRL-1 precursor (EC 3.4.21.-). | |||||
|
UROK_MOUSE
|
||||||
| NC score | 0.056986 (rank : 117) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06869 | Gene names | Plau | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase-type plasminogen activator precursor (EC 3.4.21.73) (uPA) (U-plasminogen activator) [Contains: Urokinase-type plasminogen activator long chain A; Urokinase-type plasminogen activator short chain A; Urokinase-type plasminogen activator chain B]. | |||||
|
CTRB1_HUMAN
|
||||||
| NC score | 0.056748 (rank : 118) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17538 | Gene names | CTRB1, CTRB | |||
|
Domain Architecture |

|
|||||
| Description | Chymotrypsinogen B precursor (EC 3.4.21.1) [Contains: Chymotrypsin B chain A; Chymotrypsin B chain B; Chymotrypsin B chain C]. | |||||
|
PRSS8_MOUSE
|
||||||
| NC score | 0.056619 (rank : 119) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ESD1 | Gene names | Prss8, Cap1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostasin precursor (EC 3.4.21.-) (Channel-activating protease 1) [Contains: Prostasin light chain; Prostasin heavy chain]. | |||||
|
POLS2_HUMAN
|
||||||
| NC score | 0.056547 (rank : 120) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5K4E3, Q8NBY4 | Gene names | PRSS36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
TECTB_MOUSE
|
||||||
| NC score | 0.056545 (rank : 121) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08524 | Gene names | Tectb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
TECTB_HUMAN
|
||||||
| NC score | 0.056518 (rank : 122) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PL2 | Gene names | TECTB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
ELA1_HUMAN
|
||||||
| NC score | 0.056249 (rank : 123) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNI1, Q5MLF0, Q6DJT0, Q6ISM6 | Gene names | ELA1 | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-1 precursor (EC 3.4.21.36). | |||||
|
MCPT6_MOUSE
|
||||||
| NC score | 0.056156 (rank : 124) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P21845, Q61962 | Gene names | Mcpt6 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease 6 precursor (EC 3.4.21.59) (MMCP-6) (Tryptase). | |||||
|
TGBR3_HUMAN
|
||||||
| NC score | 0.056111 (rank : 125) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03167, Q5T2T4, Q5U731, Q9UGI2 | Gene names | TGFBR3 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
ELA3A_HUMAN
|
||||||
| NC score | 0.055829 (rank : 126) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09093, Q9BRW4 | Gene names | ELA3A, ELA3 | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-3A precursor (EC 3.4.21.70) (Elastase IIIA) (Protease E). | |||||
|
PRSS8_HUMAN
|
||||||
| NC score | 0.055706 (rank : 127) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16651, Q9UCA3 | Gene names | PRSS8 | |||
|
Domain Architecture |

|
|||||
| Description | Prostasin precursor (EC 3.4.21.-) [Contains: Prostasin light chain; Prostasin heavy chain]. | |||||
|
ELA2A_HUMAN
|
||||||
| NC score | 0.055664 (rank : 128) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08217, Q14243 | Gene names | ELA2A | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-2A precursor (EC 3.4.21.71). | |||||
|
TRYA1_HUMAN
|
||||||
| NC score | 0.055606 (rank : 129) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15157, Q9H2Y5, Q9UQI1 | Gene names | TPSAB1, TPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase alpha-1 precursor (EC 3.4.21.59) (Tryptase-1). | |||||
|
TRYB1_HUMAN
|
||||||
| NC score | 0.055496 (rank : 130) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15661, Q15663, Q9H2Y4 | Gene names | TPSAB1, TPSB1 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase beta-1 precursor (EC 3.4.21.59) (Tryptase-1) (Tryptase I). | |||||
|
TRYB2_HUMAN
|
||||||
| NC score | 0.055496 (rank : 131) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20231, O95827, Q15664, Q9UQI6, Q9UQI7 | Gene names | TPSB2, TPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase beta-2 precursor (EC 3.4.21.59) (Tryptase-2) (Tryptase II). | |||||
|
ELA2B_HUMAN
|
||||||
| NC score | 0.055407 (rank : 132) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08218 | Gene names | ELA2B | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-2B precursor (EC 3.4.21.71). | |||||
|
ELA2A_MOUSE
|
||||||
| NC score | 0.055388 (rank : 133) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05208 | Gene names | Ela2a, Ela-2, Ela2 | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-2A precursor (EC 3.4.21.71) (Elastase-2). | |||||
|
ELA3B_HUMAN
|
||||||
| NC score | 0.055240 (rank : 134) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08861, P11423, Q5VU28, Q5VU29, Q5VU30 | Gene names | ELA3B | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-3B precursor (EC 3.4.21.70) (Elastase IIIB) (Protease E). | |||||
|
KREM1_HUMAN
|
||||||
| NC score | 0.055186 (rank : 135) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MU8, Q5TIB9, Q6P3X6, Q9BY70, Q9UGS5, Q9UGU1 | Gene names | KREMEN1, KREMEN, KRM1 | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
POLS2_MOUSE
|
||||||
| NC score | 0.054944 (rank : 136) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5K2P8 | Gene names | Prss36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
HGFA_HUMAN
|
||||||
| NC score | 0.054897 (rank : 137) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
PROC_HUMAN
|
||||||
| NC score | 0.054876 (rank : 138) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P04070, Q15189, Q15190, Q16001 | Gene names | PROC | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
TGBR3_MOUSE
|
||||||
| NC score | 0.054706 (rank : 139) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88393, Q6NS72 | Gene names | Tgfbr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
FA12_HUMAN
|
||||||
| NC score | 0.054402 (rank : 140) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
CFAD_MOUSE
|
||||||
| NC score | 0.054321 (rank : 141) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P03953, Q61280 | Gene names | Cfd, Adn, Df | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin) (28 kDa adipocyte protein). | |||||
|
TRYB1_MOUSE
|
||||||
| NC score | 0.054200 (rank : 142) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02844 | Gene names | Tpsab1, Mcpt7 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase precursor (EC 3.4.21.59) (Mast cell protease 7) (MMCP-7) (Tryptase alpha/beta-1). | |||||
|
FA10_HUMAN
|
||||||
| NC score | 0.054152 (rank : 143) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P00742, Q14340 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) (Stuart- Prower factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
ZP3_MOUSE
|
||||||
| NC score | 0.054065 (rank : 144) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10761 | Gene names | Zp3, Zp-3, Zpc | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Sperm receptor) (Zona pellucida protein C). | |||||
|
TRYD_HUMAN
|
||||||
| NC score | 0.053994 (rank : 145) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZJ3, O95824, Q8TDI6, Q96L36, Q96RZ5, Q9H2Y6, Q9UQI8 | Gene names | TPSD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase delta precursor (EC 3.4.21.59) (Delta tryptase) (Mast cell mMCP-7-like) (Tryptase-3) (HmMCP-3-like tryptase III). | |||||
|
FA10_MOUSE
|
||||||
| NC score | 0.053722 (rank : 146) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
FA7_MOUSE
|
||||||
| NC score | 0.053722 (rank : 147) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70375, Q61109 | Gene names | F7, Cf7 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
KLK10_HUMAN
|
||||||
| NC score | 0.053703 (rank : 148) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43240, Q99920, Q9GZW9 | Gene names | KLK10, NES1, PRRSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-10 precursor (EC 3.4.21.-) (Protease serine-like 1) (Normal epithelial cell-specific 1). | |||||
|
HGFA_MOUSE
|
||||||
| NC score | 0.053586 (rank : 149) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
CFAD_HUMAN
|
||||||
| NC score | 0.053562 (rank : 150) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00746 | Gene names | CFD, DF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin). | |||||
|
THRB_HUMAN
|
||||||
| NC score | 0.053527 (rank : 151) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00734 | Gene names | F2 | |||
|
Domain Architecture |

|
|||||
| Description | Prothrombin precursor (EC 3.4.21.5) (Coagulation factor II) [Contains: Activation peptide fragment 1; Activation peptide fragment 2; Thrombin light chain; Thrombin heavy chain]. | |||||
|
HABP2_HUMAN
|
||||||
| NC score | 0.053416 (rank : 152) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14520, O00663 | Gene names | HABP2, HGFAL, PHBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) (Hepatocyte growth factor activator-like protein) (Factor VII-activating protease) (Factor seven-activating protease) (FSAP) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
KLK6_HUMAN
|
||||||
| NC score | 0.053386 (rank : 153) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92876 | Gene names | KLK6, PRSS18, PRSS9 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-6 precursor (EC 3.4.21.-) (Protease M) (Neurosin) (Zyme) (SP59). | |||||
|
KLK13_HUMAN
|
||||||
| NC score | 0.053378 (rank : 154) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKR3, Q9Y433 | Gene names | KLK13, KLKL4 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-13 precursor (EC 3.4.21.-) (Kallikrein-like protein 4) (KLK-L4). | |||||
|
PROC_MOUSE
|
||||||
| NC score | 0.053376 (rank : 155) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
TRY3_HUMAN
|
||||||
| NC score | 0.053194 (rank : 156) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35030, P15951, Q15665, Q9UQV3 | Gene names | PRSS3, PRSS4, TRY3, TRY4 | |||
|
Domain Architecture |

|
|||||
| Description | Trypsin-3 precursor (EC 3.4.21.4) (Trypsin III) (Brain trypsinogen) (Mesotrypsinogen) (Trypsin IV). | |||||
|
PLMN_HUMAN
|
||||||
| NC score | 0.053101 (rank : 157) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00747, Q15146, Q6PA00 | Gene names | PLG | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
HABP2_MOUSE
|
||||||
| NC score | 0.053048 (rank : 158) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0D2 | Gene names | Habp2, Phbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
PLMN_MOUSE
|
||||||
| NC score | 0.052940 (rank : 159) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20918, Q8CIS2, Q91WJ5 | Gene names | Plg | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
TRY2_MOUSE
|
||||||
| NC score | 0.052908 (rank : 160) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07146 | Gene names | Prss2, Try2 | |||
|
Domain Architecture |

|
|||||
| Description | Anionic trypsin-2 precursor (EC 3.4.21.4) (Anionic trypsin II) (Pretrypsinogen II). | |||||
|
FA7_HUMAN
|
||||||
| NC score | 0.052904 (rank : 161) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08709, Q14339, Q5JVF2 | Gene names | F7 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) (SPCA) (Proconvertin) (Eptacog alfa) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
THRB_MOUSE
|
||||||
| NC score | 0.052769 (rank : 162) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19221 | Gene names | F2, Cf2 | |||
|
Domain Architecture |

|
|||||
| Description | Prothrombin precursor (EC 3.4.21.5) (Coagulation factor II) [Contains: Activation peptide fragment 1; Activation peptide fragment 2; Thrombin light chain; Thrombin heavy chain]. | |||||
|
NRPN_MOUSE
|
||||||
| NC score | 0.052563 (rank : 163) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61955 | Gene names | Klk8, Nrpn, Prss19 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8). | |||||
|
TRY1_HUMAN
|
||||||
| NC score | 0.052558 (rank : 164) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07477, Q92955, Q9HAN4, Q9HAN5, Q9HAN6, Q9HAN7 | Gene names | PRSS1, TRP1, TRY1, TRYP1 | |||
|
Domain Architecture |

|
|||||
| Description | Trypsin-1 precursor (EC 3.4.21.4) (Trypsin I) (Cationic trypsinogen). | |||||
|
APOA_HUMAN
|
||||||
| NC score | 0.052453 (rank : 165) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
TRY2_HUMAN
|
||||||
| NC score | 0.052297 (rank : 166) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07478 | Gene names | PRSS2, TRY2, TRYP2 | |||
|
Domain Architecture |

|
|||||
| Description | Trypsin-2 precursor (EC 3.4.21.4) (Trypsin II) (Anionic trypsinogen). | |||||
|
NRPN_HUMAN
|
||||||
| NC score | 0.051561 (rank : 167) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60259, Q9HCB3, Q9UIL9, Q9UQ47 | Gene names | KLK8, NRPN, PRSS19, TADG14 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8) (Ovasin) (Serine protease TADG-14) (Tumor-associated differentially expressed gene 14 protein). | |||||
|
HPT_HUMAN
|
||||||
| NC score | 0.051409 (rank : 168) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00738, P00737, Q2PP15, Q3B7J0, Q6LBY9 | Gene names | HP | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.051316 (rank : 169) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
HPTR_HUMAN
|
||||||
| NC score | 0.051294 (rank : 170) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00739, Q7LE20, Q92658, Q92659, Q9ULB0 | Gene names | HPR | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin-related protein precursor. | |||||
|
GRAK_MOUSE
|
||||||
| NC score | 0.051277 (rank : 171) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35205 | Gene names | Gzmk | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme K precursor (EC 3.4.21.-). | |||||
|
HPT_MOUSE
|
||||||
| NC score | 0.051242 (rank : 172) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61646 | Gene names | Hp | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
ZP3_HUMAN
|
||||||
| NC score | 0.050938 (rank : 173) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21754, Q06633 | Gene names | ZP3, ZP3A, ZPC | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Zona pellucida protein C) (Sperm receptor) (ZP3A/ZP3B). | |||||
|
KLK15_HUMAN
|
||||||
| NC score | 0.050814 (rank : 174) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H2R5, Q15358, Q9H2R3, Q9H2R4, Q9H2R6, Q9HBG9 | Gene names | KLK15 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-15 precursor (EC 3.4.21.-) (ACO protease). | |||||
|
KLK7_MOUSE
|
||||||
| NC score | 0.050642 (rank : 175) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91VE3, Q9R048 | Gene names | Klk7, Prss6, Scce | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kallikrein-7 precursor (EC 3.4.21.-) (Stratum corneum chymotryptic enzyme) (Thymopsin). | |||||
|
ELNE_MOUSE
|
||||||
| NC score | 0.050543 (rank : 176) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3UP87, Q61515 | Gene names | Ela2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte elastase precursor (EC 3.4.21.37) (Elastase-2) (Neutrophil elastase). | |||||
|
KLK12_HUMAN
|
||||||
| NC score | 0.050020 (rank : 177) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKR0, Q9UKR1 | Gene names | KLK12, KLKL5 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-12 precursor (EC 3.4.21.-) (Kallikrein-like protein 5) (KLK-L5). | |||||
|
UROM_HUMAN
|
||||||
| NC score | 0.035621 (rank : 178) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
XLKD1_MOUSE
|
||||||
| NC score | 0.010747 (rank : 179) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
LTB1S_MOUSE
|
||||||
| NC score | 0.007031 (rank : 180) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1L_MOUSE
|
||||||
| NC score | 0.007019 (rank : 181) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
FBLN5_MOUSE
|
||||||
| NC score | 0.003909 (rank : 182) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVH9, Q541Z7 | Gene names | Fbln5, Dance | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance). | |||||
|
ZBTB6_HUMAN
|
||||||
| NC score | -0.003620 (rank : 183) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||