Please be patient as the page loads
|
NECD_MOUSE
|
||||||
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NECD_MOUSE
|
||||||
| θ value | 4.55495e-161 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
NECD_HUMAN
|
||||||
| θ value | 1.29261e-131 (rank : 2) | NC score | 0.990127 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |
|
|||||
| Description | Necdin. | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | 2.26182e-43 (rank : 3) | NC score | 0.958777 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
TROP_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 4) | NC score | 0.919077 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
MAGG1_HUMAN
|
||||||
| θ value | 6.58091e-43 (rank : 5) | NC score | 0.966229 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGG1_MOUSE
|
||||||
| θ value | 4.2656e-42 (rank : 6) | NC score | 0.966656 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MAGF1_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 7) | NC score | 0.970945 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 3.37985e-39 (rank : 8) | NC score | 0.917421 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
MAGL2_HUMAN
|
||||||
| θ value | 1.08726e-37 (rank : 9) | NC score | 0.951393 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 10) | NC score | 0.665216 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGB4_HUMAN
|
||||||
| θ value | 5.584e-34 (rank : 11) | NC score | 0.942024 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGD1_MOUSE
|
||||||
| θ value | 5.584e-34 (rank : 12) | NC score | 0.926797 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAGD1_HUMAN
|
||||||
| θ value | 7.29293e-34 (rank : 13) | NC score | 0.937461 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MAGB1_HUMAN
|
||||||
| θ value | 1.37539e-32 (rank : 14) | NC score | 0.939507 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MAGBI_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 15) | NC score | 0.937878 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 16) | NC score | 0.929129 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MAGBA_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 17) | NC score | 0.942131 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGB3_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 18) | NC score | 0.939890 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
MAGB2_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 19) | NC score | 0.942062 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
MAGAB_HUMAN
|
||||||
| θ value | 9.2256e-29 (rank : 20) | NC score | 0.918515 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGL2_MOUSE
|
||||||
| θ value | 1.57365e-28 (rank : 21) | NC score | 0.939697 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |
|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
MAGAA_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 22) | NC score | 0.920447 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
MAGE2_HUMAN
|
||||||
| θ value | 1.02001e-27 (rank : 23) | NC score | 0.940791 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 8.63488e-27 (rank : 24) | NC score | 0.874378 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAGC2_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 25) | NC score | 0.932134 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
MAGH1_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 26) | NC score | 0.950421 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
MAGH1_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 27) | NC score | 0.949397 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
MAGA4_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 28) | NC score | 0.901750 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
MAGA9_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 29) | NC score | 0.913166 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
MAGA1_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 30) | NC score | 0.903669 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGA6_HUMAN
|
||||||
| θ value | 2.59989e-23 (rank : 31) | NC score | 0.890572 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MAGA3_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 32) | NC score | 0.887052 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |
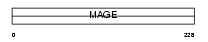
|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGAC_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 33) | NC score | 0.885939 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |
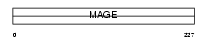
|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGA2_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 34) | NC score | 0.885048 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |
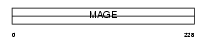
|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGC3_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 35) | NC score | 0.835595 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
MAGA8_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 36) | NC score | 0.834526 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |
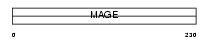
|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 37) | NC score | 0.062210 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 38) | NC score | 0.062044 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 39) | NC score | 0.061310 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 40) | NC score | 0.061661 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 41) | NC score | 0.061347 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.125558 (rank : 42) | NC score | 0.035606 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 43) | NC score | 0.065937 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 44) | NC score | 0.060072 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.058920 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.018012 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.046568 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
DACT1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.022042 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4A3, Q80VG9, Q8BP49, Q9JK89 | Gene names | Dact1, Thyex3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (MDpr1) (Frodo) (Thymus-expressed novel gene 3 protein). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.009996 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.071718 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.012032 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.032296 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CITE4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.021898 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96RK1 | Gene names | CITED4, MRG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
CSTF2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.004822 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.011973 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.024798 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.033476 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.030338 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
SYN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.022382 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.039883 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.002362 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
FXL16_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.009068 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N461, Q2MHR2, Q96S14, Q9UJI0 | Gene names | FBXL16, C16orf22, FBL16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 16 (F-box and leucine-rich repeat protein 16). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.017674 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.023564 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.025740 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
AB1IP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.022652 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.057014 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.023769 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SOS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.005381 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
SPRE3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.018281 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.030992 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.007578 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.011055 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
ESPL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.007897 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60330 | Gene names | Espl1, Esp1, Kiaa0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.015777 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | -0.003866 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.038485 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
CPT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.003490 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23786, Q5SW68, Q9BQ26 | Gene names | CPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Carnitine O-palmitoyltransferase 2, mitochondrial precursor (EC 2.3.1.21) (Carnitine palmitoyltransferase II) (CPT II). | |||||
|
KLF17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | -0.002252 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.000193 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.021758 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.004477 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.027034 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SYN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.019466 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
TMAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.013614 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZH7, Q8C6F8 | Gene names | Tmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.057280 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
GAK18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.052635 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.054975 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051310 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
GAK8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.054033 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
LORI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.076182 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
MAGA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.465205 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
PHXR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.062747 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
SELPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.053976 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
NECD_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.55495e-161 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
NECD_HUMAN
|
||||||
| NC score | 0.990127 (rank : 2) | θ value | 1.29261e-131 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99608 | Gene names | NDN | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MAGF1_HUMAN
|
||||||
| NC score | 0.970945 (rank : 3) | θ value | 4.71619e-41 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAY2, Q9H215 | Gene names | MAGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen F1 (MAGE-F1 antigen). | |||||
|
MAGG1_MOUSE
|
||||||
| NC score | 0.966656 (rank : 4) | θ value | 4.2656e-42 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CPR8, Q9D378 | Gene names | Ndnl2, Mageg1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2). | |||||
|
MAGG1_HUMAN
|
||||||
| NC score | 0.966229 (rank : 5) | θ value | 6.58091e-43 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MG7, Q8IW16, Q8TEI6, Q9H214 | Gene names | NDNL2, HCA4, MAGEG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen G1 (MAGE-G1 antigen) (Necdin-like protein 2) (Hepatocellular carcinoma-associated protein 4). | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.958777 (rank : 6) | θ value | 2.26182e-43 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MAGL2_HUMAN
|
||||||
| NC score | 0.951393 (rank : 7) | θ value | 1.08726e-37 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
MAGH1_HUMAN
|
||||||
| NC score | 0.950421 (rank : 8) | θ value | 4.28545e-26 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H213, Q5JRJ3, Q9Y5M2 | Gene names | MAGEH1, APR1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen) (Restin) (Apoptosis- related protein 1) (APR-1). | |||||
|
MAGH1_MOUSE
|
||||||
| NC score | 0.949397 (rank : 9) | θ value | 5.59698e-26 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NWG9, Q99PB0 | Gene names | Mageh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen H1 (MAGE-H1 antigen). | |||||
|
MAGBA_HUMAN
|
||||||
| NC score | 0.942131 (rank : 10) | θ value | 1.08979e-29 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96LZ2, Q494Y6, Q494Y7, Q9BZ78 | Gene names | MAGEB10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B10 (MAGE-B10 antigen). | |||||
|
MAGB2_HUMAN
|
||||||
| NC score | 0.942062 (rank : 11) | θ value | 7.06379e-29 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
MAGB4_HUMAN
|
||||||
| NC score | 0.942024 (rank : 12) | θ value | 5.584e-34 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15481, Q6FHH4, Q8IZ00 | Gene names | MAGEB4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B4 (MAGE-B4 antigen). | |||||
|
MAGE2_HUMAN
|
||||||
| NC score | 0.940791 (rank : 13) | θ value | 1.02001e-27 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD90 | Gene names | MAGEE2, HCA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E2 (MAGE-E2 antigen) (Hepatocellular carcinoma-associated protein 3). | |||||
|
MAGB3_HUMAN
|
||||||
| NC score | 0.939890 (rank : 14) | θ value | 3.17079e-29 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
MAGL2_MOUSE
|
||||||
| NC score | 0.939697 (rank : 15) | θ value | 1.57365e-28 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QZ04 | Gene names | Magel2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Protein nS7). | |||||
|
MAGB1_HUMAN
|
||||||
| NC score | 0.939507 (rank : 16) | θ value | 1.37539e-32 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43366, O00601, O75862, Q6FHJ0, Q96CW8 | Gene names | MAGEB1, MAGEL1, MAGEXP | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B1 (MAGE-B1 antigen) (MAGE-XP antigen) (DSS-AHC critical interval MAGE superfamily 10) (DAM10). | |||||
|
MAGBI_HUMAN
|
||||||
| NC score | 0.937878 (rank : 17) | θ value | 4.42448e-31 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.937461 (rank : 18) | θ value | 7.29293e-34 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MAGC2_HUMAN
|
||||||
| NC score | 0.932134 (rank : 19) | θ value | 2.51237e-26 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UBF1, Q5JZ00, Q96D45, Q9P1M6, Q9P1M7 | Gene names | MAGEC2, HCA587, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C2 (MAGE-C2 antigen) (MAGE-E1 antigen) (Hepatocellular carcinoma-associated antigen 587) (Cancer-testis antigen 10) (CT10). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.929129 (rank : 20) | θ value | 1.08979e-29 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MAGD1_MOUSE
|
||||||
| NC score | 0.926797 (rank : 21) | θ value | 5.584e-34 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
MAGAA_HUMAN
|
||||||
| NC score | 0.920447 (rank : 22) | θ value | 2.05525e-28 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
TROP_HUMAN
|
||||||
| NC score | 0.919077 (rank : 23) | θ value | 3.85808e-43 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12816, Q9NU89, Q9UPN8 | Gene names | TRO, KIAA1114, MAGED3 | |||
|
Domain Architecture |

|
|||||
| Description | Trophinin (MAGE-D3 antigen). | |||||
|
MAGAB_HUMAN
|
||||||
| NC score | 0.918515 (rank : 24) | θ value | 9.2256e-29 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43364, Q5ETU4, Q6ZRZ5 | Gene names | MAGEA11, MAGE11 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 11 (MAGE-11 antigen). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.917421 (rank : 25) | θ value | 3.37985e-39 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
MAGA9_HUMAN
|
||||||
| NC score | 0.913166 (rank : 26) | θ value | 1.05554e-24 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43362, Q92910 | Gene names | MAGEA9, MAGE9 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 9 (MAGE-9 antigen). | |||||
|
MAGA1_HUMAN
|
||||||
| NC score | 0.903669 (rank : 27) | θ value | 6.84181e-24 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43355, O00346 | Gene names | MAGEA1, MAGE1, MAGE1A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 1 (MAGE-1 antigen) (Antigen MZ2-E). | |||||
|
MAGA4_HUMAN
|
||||||
| NC score | 0.901750 (rank : 28) | θ value | 1.62847e-25 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43358, Q14798 | Gene names | MAGEA4, MAGE4 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 4 (MAGE-4 antigen) (MAGE-X2) (MAGE-41). | |||||
|
MAGA6_HUMAN
|
||||||
| NC score | 0.890572 (rank : 29) | θ value | 2.59989e-23 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43360 | Gene names | MAGEA6, MAGE6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 6 (MAGE-6 antigen) (MAGE3B). | |||||
|
MAGA3_HUMAN
|
||||||
| NC score | 0.887052 (rank : 30) | θ value | 2.69047e-20 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43357 | Gene names | MAGEA3, MAGE3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 3 (MAGE-3 antigen) (Antigen MZ2-D). | |||||
|
MAGAC_HUMAN
|
||||||
| NC score | 0.885939 (rank : 31) | θ value | 1.33526e-19 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
MAGA2_HUMAN
|
||||||
| NC score | 0.885048 (rank : 32) | θ value | 7.32683e-18 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43356 | Gene names | MAGEA2, MAGE2, MAGEA2A | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 2 (MAGE-2 antigen). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.874378 (rank : 33) | θ value | 8.63488e-27 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAGC3_HUMAN
|
||||||
| NC score | 0.835595 (rank : 34) | θ value | 1.80886e-08 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
MAGA8_HUMAN
|
||||||
| NC score | 0.834526 (rank : 35) | θ value | 4.92598e-06 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.665216 (rank : 36) | θ value | 2.96089e-35 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAGA5_HUMAN
|
||||||
| NC score | 0.465205 (rank : 37) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.076182 (rank : 38) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.071718 (rank : 39) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.065937 (rank : 40) | θ value | 0.125558 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
PHXR5_MOUSE
|
||||||
| NC score | 0.062747 (rank : 41) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.062210 (rank : 42) | θ value | 0.0148317 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.062044 (rank : 43) | θ value | 0.0330416 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.061661 (rank : 44) | θ value | 0.0431538 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.061347 (rank : 45) | θ value | 0.0431538 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.061310 (rank : 46) | θ value | 0.0431538 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.060072 (rank : 47) | θ value | 0.163984 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.058920 (rank : 48) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.057280 (rank : 49) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.057014 (rank : 50) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.054975 (rank : 51) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.054033 (rank : 52) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.053976 (rank : 53) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.052635 (rank : 54) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK7_HUMAN
|
||||||
| NC score | 0.051310 (rank : 55) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.046568 (rank : 56) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.039883 (rank : 57) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.038485 (rank : 58) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.035606 (rank : 59) | θ value | 0.125558 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.033476 (rank : 60) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.032296 (rank : 61) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.030992 (rank : 62) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.030338 (rank : 63) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.027034 (rank : 64) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.025740 (rank : 65) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.024798 (rank : 66) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.023769 (rank : 67) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.023564 (rank : 68) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
AB1IP_HUMAN
|
||||||
| NC score | 0.022652 (rank : 69) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
SYN2_HUMAN
|
||||||
| NC score | 0.022382 (rank : 70) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
DACT1_MOUSE
|
||||||
| NC score | 0.022042 (rank : 71) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4A3, Q80VG9, Q8BP49, Q9JK89 | Gene names | Dact1, Thyex3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (MDpr1) (Frodo) (Thymus-expressed novel gene 3 protein). | |||||
|
CITE4_HUMAN
|
||||||
| NC score | 0.021898 (rank : 72) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96RK1 | Gene names | CITED4, MRG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.021758 (rank : 73) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SYN2_MOUSE
|
||||||
| NC score | 0.019466 (rank : 74) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
SPRE3_MOUSE
|
||||||
| NC score | 0.018281 (rank : 75) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.018012 (rank : 76) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.017674 (rank : 77) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.015777 (rank : 78) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
TMAP1_MOUSE
|
||||||
| NC score | 0.013614 (rank : 79) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZH7, Q8C6F8 | Gene names | Tmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.012032 (rank : 80) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.011973 (rank : 81) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.011055 (rank : 82) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.009996 (rank : 83) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
FXL16_HUMAN
|
||||||
| NC score | 0.009068 (rank : 84) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N461, Q2MHR2, Q96S14, Q9UJI0 | Gene names | FBXL16, C16orf22, FBL16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 16 (F-box and leucine-rich repeat protein 16). | |||||
|
ESPL1_MOUSE
|
||||||
| NC score | 0.007897 (rank : 85) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60330 | Gene names | Espl1, Esp1, Kiaa0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.007578 (rank : 86) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
SOS2_HUMAN
|
||||||
| NC score | 0.005381 (rank : 87) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07890, Q15503 | Gene names | SOS2 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 2 (SOS-2). | |||||
|
CSTF2_MOUSE
|
||||||
| NC score | 0.004822 (rank : 88) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.004477 (rank : 89) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
CPT2_HUMAN
|
||||||
| NC score | 0.003490 (rank : 90) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23786, Q5SW68, Q9BQ26 | Gene names | CPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Carnitine O-palmitoyltransferase 2, mitochondrial precursor (EC 2.3.1.21) (Carnitine palmitoyltransferase II) (CPT II). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.002362 (rank : 91) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
PCD15_HUMAN
|
||||||
| NC score | 0.000193 (rank : 92) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
KLF17_HUMAN
|
||||||
| NC score | -0.002252 (rank : 93) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | -0.003866 (rank : 94) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||