Please be patient as the page loads
|
STAG2_HUMAN
|
||||||
| SwissProt Accessions | Q8N3U4, O00540, Q5JTI6, Q9H1N8 | Gene names | STAG2, SA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STAG1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993341 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
STAG1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993841 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D3E6, O08982 | Gene names | Stag1, Sa1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
STAG2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N3U4, O00540, Q5JTI6, Q9H1N8 | Gene names | STAG2, SA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
STAG2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999474 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35638, Q6NZN7 | Gene names | Stag2, Sa2, Sap2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
STAG3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.983441 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJ98, Q8NDP3 | Gene names | STAG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
STAG3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.984214 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70576 | Gene names | Stag3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.026499 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.017427 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ZWINT_MOUSE
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.031420 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
CJ068_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.068827 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
ASCC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.032465 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WR3, Q8BKM9, Q8BX60, Q8R1B9 | Gene names | Ascc2, Asc1p100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 2 (ASC-1 complex subunit p100) (Trip4 complex subunit p100). | |||||
|
SAS10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.066122 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JI13, Q3UL95, Q8R5C6, Q9JJ12 | Gene names | Crlz1, Crl1, Sas10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1) (Crl-1). | |||||
|
KLC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.017049 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
MCM5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.013914 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.007900 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
NCKX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.025055 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UI40, Q9NTN5, Q9NZQ4 | Gene names | SLC24A2, NCKX2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 2 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 2) (Retinal cone Na-Ca+K exchanger) (Solute carrier family 24 member 2). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.021465 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
PIWL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.016026 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMB7 | Gene names | Piwil1, Miwi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
OMGP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.008484 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63912 | Gene names | Omg, Omgp | |||
|
Domain Architecture |
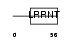
|
|||||
| Description | Oligodendrocyte-myelin glycoprotein precursor. | |||||
|
IQCB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.017132 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BP00, Q3TNK4, Q8K306 | Gene names | Iqcb1, Kiaa0036 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ calmodulin-binding motif-containing protein 1. | |||||
|
PIWL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.013847 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J94, O95404, Q8NA60, Q8TBY5, Q96JD5 | Gene names | PIWIL1, HIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
SAS10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.046533 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
CCD93_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.007442 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
PEX5R_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.013466 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.008759 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CRCM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.007564 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |
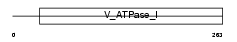
|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
HERC5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.005642 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
KAD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.010004 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUR9, Q9R1X7 | Gene names | Ak3l1, Ak-4, Ak3b, Ak4 | |||
|
Domain Architecture |
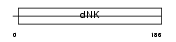
|
|||||
| Description | Adenylate kinase isoenzyme 4, mitochondrial (EC 2.7.4.3) (Adenylate kinase 3-like 1) (ATP-AMP transphosphorylase). | |||||
|
NRDC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.015693 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
PRAM7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.006002 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VXH5 | Gene names | PRAMEF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 7. | |||||
|
PRAM8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.005999 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VWM4 | Gene names | PRAMEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 8. | |||||
|
STAG2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N3U4, O00540, Q5JTI6, Q9H1N8 | Gene names | STAG2, SA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
STAG2_MOUSE
|
||||||
| NC score | 0.999474 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35638, Q6NZN7 | Gene names | Stag2, Sa2, Sap2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
STAG1_MOUSE
|
||||||
| NC score | 0.993841 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D3E6, O08982 | Gene names | Stag1, Sa1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
STAG1_HUMAN
|
||||||
| NC score | 0.993341 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
STAG3_MOUSE
|
||||||
| NC score | 0.984214 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70576 | Gene names | Stag3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
STAG3_HUMAN
|
||||||
| NC score | 0.983441 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJ98, Q8NDP3 | Gene names | STAG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
CJ068_HUMAN
|
||||||
| NC score | 0.068827 (rank : 7) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
SAS10_MOUSE
|
||||||
| NC score | 0.066122 (rank : 8) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JI13, Q3UL95, Q8R5C6, Q9JJ12 | Gene names | Crlz1, Crl1, Sas10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1) (Crl-1). | |||||
|
SAS10_HUMAN
|
||||||
| NC score | 0.046533 (rank : 9) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
ASCC2_MOUSE
|
||||||
| NC score | 0.032465 (rank : 10) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WR3, Q8BKM9, Q8BX60, Q8R1B9 | Gene names | Ascc2, Asc1p100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 2 (ASC-1 complex subunit p100) (Trip4 complex subunit p100). | |||||
|
ZWINT_MOUSE
|
||||||
| NC score | 0.031420 (rank : 11) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.026499 (rank : 12) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
NCKX2_HUMAN
|
||||||
| NC score | 0.025055 (rank : 13) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UI40, Q9NTN5, Q9NZQ4 | Gene names | SLC24A2, NCKX2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 2 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 2) (Retinal cone Na-Ca+K exchanger) (Solute carrier family 24 member 2). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.021465 (rank : 14) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.017427 (rank : 15) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
IQCB1_MOUSE
|
||||||
| NC score | 0.017132 (rank : 16) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BP00, Q3TNK4, Q8K306 | Gene names | Iqcb1, Kiaa0036 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ calmodulin-binding motif-containing protein 1. | |||||
|
KLC2_MOUSE
|
||||||
| NC score | 0.017049 (rank : 17) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
PIWL1_MOUSE
|
||||||
| NC score | 0.016026 (rank : 18) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMB7 | Gene names | Piwil1, Miwi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
NRDC_HUMAN
|
||||||
| NC score | 0.015693 (rank : 19) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
MCM5_HUMAN
|
||||||
| NC score | 0.013914 (rank : 20) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
PIWL1_HUMAN
|
||||||
| NC score | 0.013847 (rank : 21) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J94, O95404, Q8NA60, Q8TBY5, Q96JD5 | Gene names | PIWIL1, HIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
PEX5R_HUMAN
|
||||||
| NC score | 0.013466 (rank : 22) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
KAD4_MOUSE
|
||||||
| NC score | 0.010004 (rank : 23) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUR9, Q9R1X7 | Gene names | Ak3l1, Ak-4, Ak3b, Ak4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 4, mitochondrial (EC 2.7.4.3) (Adenylate kinase 3-like 1) (ATP-AMP transphosphorylase). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.008759 (rank : 24) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
OMGP_MOUSE
|
||||||
| NC score | 0.008484 (rank : 25) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63912 | Gene names | Omg, Omgp | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte-myelin glycoprotein precursor. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.007900 (rank : 26) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CRCM_HUMAN
|
||||||
| NC score | 0.007564 (rank : 27) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |

|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
CCD93_HUMAN
|
||||||
| NC score | 0.007442 (rank : 28) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
PRAM7_HUMAN
|
||||||
| NC score | 0.006002 (rank : 29) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VXH5 | Gene names | PRAMEF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 7. | |||||
|
PRAM8_HUMAN
|
||||||
| NC score | 0.005999 (rank : 30) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VWM4 | Gene names | PRAMEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member 8. | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.005642 (rank : 31) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||