Please be patient as the page loads
|
NRDC_HUMAN
|
||||||
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NRDC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.986043 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_HUMAN
|
||||||
| θ value | 5.22361e-149 (rank : 3) | NC score | 0.920153 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_MOUSE
|
||||||
| θ value | 5.22361e-149 (rank : 4) | NC score | 0.920621 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
MPPA_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 5) | NC score | 0.273791 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPB_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 6) | NC score | 0.262346 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPB_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 7) | NC score | 0.246865 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
UQCR1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 8) | NC score | 0.246734 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 9) | NC score | 0.261086 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |
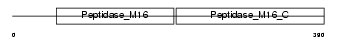
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 10) | NC score | 0.102695 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 11) | NC score | 0.033480 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
MPPA_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 12) | NC score | 0.241957 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |
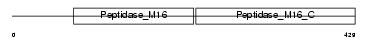
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.096172 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.014112 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.085370 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.090106 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TINAL_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.064217 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
PREP_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.128089 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K411, Q3THC4, Q3TJI1, Q3TPP0, Q3URQ8, Q4KUG2, Q4KUG3, Q6ZPX6, Q922N1, Q9CV63 | Gene names | Pitrm1, Kiaa1104, Ntup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (Pitrilysin metalloproteinase 1). | |||||
|
PREP_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.134074 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JRX3, O95204, Q2M2G6, Q4VBR1, Q5JRW7, Q7L5Z7, Q9BSI6, Q9BVJ5, Q9UPP8 | Gene names | PITRM1, KIAA1104, MP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (hPreP) (Pitrilysin metalloproteinase 1) (Metalloprotease 1) (hMP1). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.068110 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.078092 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TINAL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.055768 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.058177 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.033071 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.029452 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.016911 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.081411 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.066058 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NC6IP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.040653 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.020166 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
SH3G1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.020410 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62419 | Gene names | Sh3gl1, Sh3d2b | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (SH3p8). | |||||
|
APOB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.026343 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04114, O00502, P78479, P78480, P78481, Q13779, Q13785, Q13786, Q13787, Q13788, Q7Z600, Q9UMN0 | Gene names | APOB | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein B-100 precursor (Apo B-100) [Contains: Apolipoprotein B-48 (Apo B-48)]. | |||||
|
CCD82_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.049996 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
SH3G1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.019054 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99961, Q99668 | Gene names | SH3GL1, CNSA1, SH3D2B | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (Extra eleven-nineteen leukemia fusion gene) (EEN) (EEN fusion partner of MLL). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.040611 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TF2AY_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.027417 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.015045 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.015552 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
HDGR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.030888 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
RAG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.034670 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.028312 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
CASC5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.016985 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.034465 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
TPA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.002436 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11214, Q91VP2 | Gene names | Plat | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.012104 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
GPTC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.046812 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.035868 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
SUCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.028375 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUM5, Q8C2C3, Q9DBA3, Q9DCI8 | Gene names | Suclg1 | |||
|
Domain Architecture |
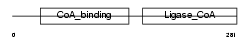
|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.035070 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.034591 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.004116 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.004505 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
MEPE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.018003 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |
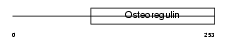
|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
SEPT8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.009592 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92599, Q8IX36, Q8IX37, Q9BVB3 | Gene names | SEPT8, KIAA0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
STAG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.015693 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3U4, O00540, Q5JTI6, Q9H1N8 | Gene names | STAG2, SA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
STAG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.015713 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35638, Q6NZN7 | Gene names | Stag2, Sa2, Sap2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
STX6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.010853 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43752, Q5VY08, Q6FH83 | Gene names | STX6 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-6. | |||||
|
SUCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.025279 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53597, Q9BWB0, Q9UNP6 | Gene names | SUCLG1 | |||
|
Domain Architecture |
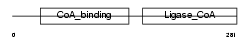
|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.016818 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TTLL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.013226 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |
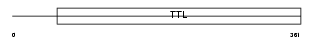
|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.067356 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.065809 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.053159 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
UQCR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.089733 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |
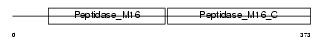
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.091385 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |
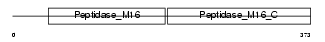
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
NRDC_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.986043 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_MOUSE
|
||||||
| NC score | 0.920621 (rank : 3) | θ value | 5.22361e-149 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| NC score | 0.920153 (rank : 4) | θ value | 5.22361e-149 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
MPPA_MOUSE
|
||||||
| NC score | 0.273791 (rank : 5) | θ value | 4.1701e-05 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPB_HUMAN
|
||||||
| NC score | 0.262346 (rank : 6) | θ value | 9.29e-05 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
UQCR1_HUMAN
|
||||||
| NC score | 0.261086 (rank : 7) | θ value | 0.000461057 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPB_MOUSE
|
||||||
| NC score | 0.246865 (rank : 8) | θ value | 0.000270298 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
UQCR1_MOUSE
|
||||||
| NC score | 0.246734 (rank : 9) | θ value | 0.000270298 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPA_HUMAN
|
||||||
| NC score | 0.241957 (rank : 10) | θ value | 0.00390308 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
PREP_HUMAN
|
||||||
| NC score | 0.134074 (rank : 11) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JRX3, O95204, Q2M2G6, Q4VBR1, Q5JRW7, Q7L5Z7, Q9BSI6, Q9BVJ5, Q9UPP8 | Gene names | PITRM1, KIAA1104, MP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (hPreP) (Pitrilysin metalloproteinase 1) (Metalloprotease 1) (hMP1). | |||||
|
PREP_MOUSE
|
||||||
| NC score | 0.128089 (rank : 12) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K411, Q3THC4, Q3TJI1, Q3TPP0, Q3URQ8, Q4KUG2, Q4KUG3, Q6ZPX6, Q922N1, Q9CV63 | Gene names | Pitrm1, Kiaa1104, Ntup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (Pitrilysin metalloproteinase 1). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.102695 (rank : 13) | θ value | 0.00175202 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.096172 (rank : 14) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
UQCR2_MOUSE
|
||||||
| NC score | 0.091385 (rank : 15) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.090106 (rank : 16) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
UQCR2_HUMAN
|
||||||
| NC score | 0.089733 (rank : 17) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.085370 (rank : 18) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.081411 (rank : 19) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.078092 (rank : 20) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.068110 (rank : 21) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.067356 (rank : 22) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.066058 (rank : 23) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.065809 (rank : 24) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
TINAL_MOUSE
|
||||||
| NC score | 0.064217 (rank : 25) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.058177 (rank : 26) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TINAL_HUMAN
|
||||||
| NC score | 0.055768 (rank : 27) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.053159 (rank : 28) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
CCD82_HUMAN
|
||||||
| NC score | 0.049996 (rank : 29) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
GPTC3_HUMAN
|
||||||
| NC score | 0.046812 (rank : 30) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
NC6IP_MOUSE
|
||||||
| NC score | 0.040653 (rank : 31) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.040611 (rank : 32) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.035868 (rank : 33) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.035070 (rank : 34) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
RAG2_MOUSE
|
||||||
| NC score | 0.034670 (rank : 35) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.034591 (rank : 36) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.034465 (rank : 37) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.033480 (rank : 38) | θ value | 0.00228821 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.033071 (rank : 39) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
HDGR3_HUMAN
|
||||||
| NC score | 0.030888 (rank : 40) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.029452 (rank : 41) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SUCA_MOUSE
|
||||||
| NC score | 0.028375 (rank : 42) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUM5, Q8C2C3, Q9DBA3, Q9DCI8 | Gene names | Suclg1 | |||
|
Domain Architecture |

|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.028312 (rank : 43) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
TF2AY_MOUSE
|
||||||
| NC score | 0.027417 (rank : 44) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
APOB_HUMAN
|
||||||
| NC score | 0.026343 (rank : 45) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04114, O00502, P78479, P78480, P78481, Q13779, Q13785, Q13786, Q13787, Q13788, Q7Z600, Q9UMN0 | Gene names | APOB | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein B-100 precursor (Apo B-100) [Contains: Apolipoprotein B-48 (Apo B-48)]. | |||||
|
SUCA_HUMAN
|
||||||
| NC score | 0.025279 (rank : 46) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53597, Q9BWB0, Q9UNP6 | Gene names | SUCLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
SH3G1_MOUSE
|
||||||
| NC score | 0.020410 (rank : 47) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62419 | Gene names | Sh3gl1, Sh3d2b | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (SH3p8). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.020166 (rank : 48) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
SH3G1_HUMAN
|
||||||
| NC score | 0.019054 (rank : 49) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99961, Q99668 | Gene names | SH3GL1, CNSA1, SH3D2B | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (Extra eleven-nineteen leukemia fusion gene) (EEN) (EEN fusion partner of MLL). | |||||
|
MEPE_HUMAN
|
||||||
| NC score | 0.018003 (rank : 50) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |

|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
CASC5_HUMAN
|
||||||
| NC score | 0.016985 (rank : 51) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.016911 (rank : 52) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.016818 (rank : 53) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
STAG2_MOUSE
|
||||||
| NC score | 0.015713 (rank : 54) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35638, Q6NZN7 | Gene names | Stag2, Sa2, Sap2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
STAG2_HUMAN
|
||||||
| NC score | 0.015693 (rank : 55) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3U4, O00540, Q5JTI6, Q9H1N8 | Gene names | STAG2, SA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.015552 (rank : 56) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.015045 (rank : 57) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.014112 (rank : 58) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
TTLL1_HUMAN
|
||||||
| NC score | 0.013226 (rank : 59) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |

|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.012104 (rank : 60) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
STX6_HUMAN
|
||||||
| NC score | 0.010853 (rank : 61) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43752, Q5VY08, Q6FH83 | Gene names | STX6 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-6. | |||||
|
SEPT8_HUMAN
|
||||||
| NC score | 0.009592 (rank : 62) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92599, Q8IX36, Q8IX37, Q9BVB3 | Gene names | SEPT8, KIAA0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.004505 (rank : 63) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.004116 (rank : 64) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
TPA_MOUSE
|
||||||
| NC score | 0.002436 (rank : 65) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11214, Q91VP2 | Gene names | Plat | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||