Please be patient as the page loads
|
IDE_MOUSE
|
||||||
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IDE_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998956 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 2.26584e-152 (rank : 3) | NC score | 0.911024 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_HUMAN
|
||||||
| θ value | 5.22361e-149 (rank : 4) | NC score | 0.920621 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
UQCR1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 5) | NC score | 0.253612 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPB_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 6) | NC score | 0.253508 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 7) | NC score | 0.262144 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPA_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 8) | NC score | 0.234860 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
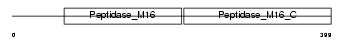
MPPB_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 9) | NC score | 0.234758 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
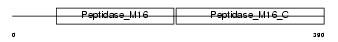
UQCR1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.230003 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
PREP_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.135856 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JRX3, O95204, Q2M2G6, Q4VBR1, Q5JRW7, Q7L5Z7, Q9BSI6, Q9BVJ5, Q9UPP8 | Gene names | PITRM1, KIAA1104, MP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (hPreP) (Pitrilysin metalloproteinase 1) (Metalloprotease 1) (hMP1). | |||||
|
PREP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.123844 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K411, Q3THC4, Q3TJI1, Q3TPP0, Q3URQ8, Q4KUG2, Q4KUG3, Q6ZPX6, Q922N1, Q9CV63 | Gene names | Pitrm1, Kiaa1104, Ntup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (Pitrilysin metalloproteinase 1). | |||||
|
PCDGA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.005813 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
4EBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.019639 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60876, Q3TDS8, Q9CZ40 | Gene names | Eif4ebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) (eIF4E-binding protein 1) (Phosphorylated heat- and acid-stable protein regulated by insulin 1) (PHAS-I). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.011704 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.018483 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
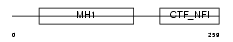
NFIB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.009624 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |
|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NFIB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.009554 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97863, P70252, P70253, P70254, Q9R1G4 | Gene names | Nfib | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
PRI1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.012041 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20664 | Gene names | Prim1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase small subunit (EC 2.7.7.-) (DNA primase 49 kDa subunit) (p49). | |||||
|
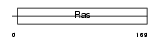
RAB30_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.003722 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15771, Q6FGK1, Q96CI8 | Gene names | RAB30 | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-30. | |||||
|
RAB30_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.003722 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923S9, Q3UQR3, Q9D3Q6 | Gene names | Rab30, Rsb30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-30. | |||||
|
TTK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.002115 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33981 | Gene names | TTK, MPS1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (Phosphotyrosine picked threonine-protein kinase) (PYT). | |||||
|
UQCR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.083166 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.084719 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
IDE_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| NC score | 0.998956 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
NRDC_HUMAN
|
||||||
| NC score | 0.920621 (rank : 3) | θ value | 5.22361e-149 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.911024 (rank : 4) | θ value | 2.26584e-152 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
MPPA_MOUSE
|
||||||
| NC score | 0.262144 (rank : 5) | θ value | 0.000602161 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
UQCR1_HUMAN
|
||||||
| NC score | 0.253612 (rank : 6) | θ value | 0.000121331 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPB_HUMAN
|
||||||
| NC score | 0.253508 (rank : 7) | θ value | 0.000158464 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_HUMAN
|
||||||
| NC score | 0.234860 (rank : 8) | θ value | 0.00228821 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPB_MOUSE
|
||||||
| NC score | 0.234758 (rank : 9) | θ value | 0.00228821 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
UQCR1_MOUSE
|
||||||
| NC score | 0.230003 (rank : 10) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
PREP_HUMAN
|
||||||
| NC score | 0.135856 (rank : 11) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JRX3, O95204, Q2M2G6, Q4VBR1, Q5JRW7, Q7L5Z7, Q9BSI6, Q9BVJ5, Q9UPP8 | Gene names | PITRM1, KIAA1104, MP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (hPreP) (Pitrilysin metalloproteinase 1) (Metalloprotease 1) (hMP1). | |||||
|
PREP_MOUSE
|
||||||
| NC score | 0.123844 (rank : 12) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K411, Q3THC4, Q3TJI1, Q3TPP0, Q3URQ8, Q4KUG2, Q4KUG3, Q6ZPX6, Q922N1, Q9CV63 | Gene names | Pitrm1, Kiaa1104, Ntup1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Presequence protease, mitochondrial precursor (EC 3.4.24.-) (Pitrilysin metalloproteinase 1). | |||||
|
UQCR2_MOUSE
|
||||||
| NC score | 0.084719 (rank : 13) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_HUMAN
|
||||||
| NC score | 0.083166 (rank : 14) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
4EBP1_MOUSE
|
||||||
| NC score | 0.019639 (rank : 15) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60876, Q3TDS8, Q9CZ40 | Gene names | Eif4ebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) (eIF4E-binding protein 1) (Phosphorylated heat- and acid-stable protein regulated by insulin 1) (PHAS-I). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.018483 (rank : 16) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
PRI1_MOUSE
|
||||||
| NC score | 0.012041 (rank : 17) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20664 | Gene names | Prim1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase small subunit (EC 2.7.7.-) (DNA primase 49 kDa subunit) (p49). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.011704 (rank : 18) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NFIB_HUMAN
|
||||||
| NC score | 0.009624 (rank : 19) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NFIB_MOUSE
|
||||||
| NC score | 0.009554 (rank : 20) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97863, P70252, P70253, P70254, Q9R1G4 | Gene names | Nfib | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
PCDGA_HUMAN
|
||||||
| NC score | 0.005813 (rank : 21) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
RAB30_HUMAN
|
||||||
| NC score | 0.003722 (rank : 22) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15771, Q6FGK1, Q96CI8 | Gene names | RAB30 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-30. | |||||
|
RAB30_MOUSE
|
||||||
| NC score | 0.003722 (rank : 23) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923S9, Q3UQR3, Q9D3Q6 | Gene names | Rab30, Rsb30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-30. | |||||
|
TTK_HUMAN
|
||||||
| NC score | 0.002115 (rank : 24) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33981 | Gene names | TTK, MPS1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (Phosphotyrosine picked threonine-protein kinase) (PYT). | |||||