Please be patient as the page loads
|
LNP_MOUSE
|
||||||
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LNP_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
LNP_HUMAN
|
||||||
| θ value | 7.01099e-170 (rank : 2) | NC score | 0.971695 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9C0E8, Q2M2V8, Q2YD99, Q658W8, Q8N5V9, Q96MS5 | Gene names | LNP, KIAA1715 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark. | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 3) | NC score | 0.128072 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 4) | NC score | 0.095017 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 5) | NC score | 0.126583 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.126348 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.126674 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.048724 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.125607 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
AMOL2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.039943 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
SEM6B_MOUSE
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.023096 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
HEMGN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.069742 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXL5, Q6XAR3, Q86XY5, Q9NPC0 | Gene names | HEMGN, EDAG, NDR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemogen (Hemopoietic gene protein) (Negative differentiation regulator protein) (Erythroid differentiation-associated gene protein) (EDAG-1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.052432 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.047058 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.069009 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
SEN34_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.068684 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.069011 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.120192 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.016854 (rank : 92) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.041188 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.118568 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.065082 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ZFP13_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.003497 (rank : 107) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.052629 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.055814 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
FLCN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.059720 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFG4, Q6ZRX1, Q96BD2, Q96BE4 | Gene names | FLCN, BHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Folliculin (Birt-Hogg-Dube syndrome protein) (BHD skin lesion fibrofolliculoma protein). | |||||
|
HDAC6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.027761 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
LRRC7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.016657 (rank : 93) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TE7 | Gene names | Lrrc7, Kiaa1365, Lap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.051618 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.047602 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.038261 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CN103_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.043185 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96BY7, Q6ZRE7, Q9NW80 | Gene names | C14orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf103. | |||||
|
FLCN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.056225 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZS3, Q3U4U8, Q3UFZ1, Q5SWZ2, Q5SX01, Q5SX02, Q8CAC0 | Gene names | Flcn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Folliculin. | |||||
|
ICB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.038714 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YX0 | Gene names | Icb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein homolog (ICB-1 protein). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.025604 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
SPAG4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.034916 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NPE6, O43648 | Gene names | SPAG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 4 protein (Outer dense fiber-associated protein SPAG4). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.020404 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.030194 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.042233 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
GAK7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.112961 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.032726 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CING_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.015348 (rank : 96) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CING_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.015230 (rank : 97) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
GP156_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.027011 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.025985 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.025448 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PP16A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.008800 (rank : 103) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |
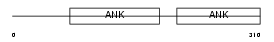
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.031047 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.026336 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.058983 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAD12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.003798 (rank : 106) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55289 | Gene names | CDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-12 precursor (Brain-cadherin) (BR-cadherin) (N-cadherin 2) (Neural type cadherin 2). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.043420 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
GNL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.020968 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.012642 (rank : 100) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.029071 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.004833 (rank : 105) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
MTG8R_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.018819 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.025557 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.018095 (rank : 91) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
SPAG4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.030121 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJF2 | Gene names | Spag4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 4 protein (Outer dense fiber-associated protein SPAG4). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.009953 (rank : 101) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SRC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.000629 (rank : 111) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05480 | Gene names | Src | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal proto-oncogene tyrosine-protein kinase Src (EC 2.7.10.2) (p60-Src) (c-Src) (pp60c-src). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.016311 (rank : 94) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
CN103_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.037234 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80XK6, Q8R1H5, Q9CWK6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf103 homolog. | |||||
|
INP5E_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.028516 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.024451 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.021377 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.020929 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.031297 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
XKR7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.021217 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
ZN710_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.001855 (rank : 109) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
ANR25_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.009835 (rank : 102) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.008137 (rank : 104) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.015435 (rank : 95) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.013192 (rank : 98) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
LRRC7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.013103 (rank : 99) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.030228 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.002857 (rank : 108) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.001618 (rank : 110) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
SON_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.033344 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.031121 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TDRD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.019819 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
ZNHI2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.027092 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY66, Q8C356, Q8VCQ7 | Gene names | Znhit2, ORF6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger HIT domain-containing protein 2 (Protein FON). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.054101 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
GAK13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.083961 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62686 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1p13.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.086950 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62687 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_16p3.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.093640 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
GAK17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.094150 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.099996 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.090255 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.102377 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.102034 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.099785 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.094045 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
IGEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.060083 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P03975 | Gene names | Iap | |||
|
Domain Architecture |

|
|||||
| Description | IgE-binding protein. | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.065069 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.069001 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.067568 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.067737 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.059128 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.056807 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.066458 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.052443 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.054547 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050434 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.056265 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.058402 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.057614 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.053693 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.053985 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.061408 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
LNP_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
LNP_HUMAN
|
||||||
| NC score | 0.971695 (rank : 2) | θ value | 7.01099e-170 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9C0E8, Q2M2V8, Q2YD99, Q658W8, Q8N5V9, Q96MS5 | Gene names | LNP, KIAA1715 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.128072 (rank : 3) | θ value | 0.0563607 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.126674 (rank : 4) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.126583 (rank : 5) | θ value | 0.0961366 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.126348 (rank : 6) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.125607 (rank : 7) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.120192 (rank : 8) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.118568 (rank : 9) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK7_HUMAN
|
||||||
| NC score | 0.112961 (rank : 10) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.102377 (rank : 11) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.102034 (rank : 12) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.099996 (rank : 13) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.099785 (rank : 14) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.095017 (rank : 15) | θ value | 0.0563607 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
GAK17_HUMAN
|
||||||
| NC score | 0.094150 (rank : 16) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK9_HUMAN
|
||||||
| NC score | 0.094045 (rank : 17) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
GAK15_HUMAN
|
||||||
| NC score | 0.093640 (rank : 18) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
GAK19_HUMAN
|
||||||
| NC score | 0.090255 (rank : 19) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK14_HUMAN
|
||||||
| NC score | 0.086950 (rank : 20) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62687 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_16p3.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK13_HUMAN
|
||||||
| NC score | 0.083961 (rank : 21) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62686 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1p13.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
HEMGN_HUMAN
|
||||||
| NC score | 0.069742 (rank : 22) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXL5, Q6XAR3, Q86XY5, Q9NPC0 | Gene names | HEMGN, EDAG, NDR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemogen (Hemopoietic gene protein) (Negative differentiation regulator protein) (Erythroid differentiation-associated gene protein) (EDAG-1). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.069011 (rank : 23) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.069009 (rank : 24) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.069001 (rank : 25) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SEN34_MOUSE
|
||||||
| NC score | 0.068684 (rank : 26) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.067737 (rank : 27) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.067568 (rank : 28) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.066458 (rank : 29) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.065082 (rank : 30) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.065069 (rank : 31) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.061408 (rank : 32) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
IGEB_MOUSE
|
||||||
| NC score | 0.060083 (rank : 33) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P03975 | Gene names | Iap | |||
|
Domain Architecture |

|
|||||
| Description | IgE-binding protein. | |||||
|
FLCN_HUMAN
|
||||||
| NC score | 0.059720 (rank : 34) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFG4, Q6ZRX1, Q96BD2, Q96BE4 | Gene names | FLCN, BHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Folliculin (Birt-Hogg-Dube syndrome protein) (BHD skin lesion fibrofolliculoma protein). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.059128 (rank : 35) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.058983 (rank : 36) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.058402 (rank : 37) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.057614 (rank : 38) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.056807 (rank : 39) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.056265 (rank : 40) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
FLCN_MOUSE
|
||||||
| NC score | 0.056225 (rank : 41) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZS3, Q3U4U8, Q3UFZ1, Q5SWZ2, Q5SX01, Q5SX02, Q8CAC0 | Gene names | Flcn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Folliculin. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.055814 (rank : 42) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.054547 (rank : 43) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.054101 (rank : 44) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.053985 (rank : 45) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.053693 (rank : 46) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.052629 (rank : 47) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.052443 (rank : 48) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.052432 (rank : 49) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.051618 (rank : 50) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.050434 (rank : 51) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.048724 (rank : 52) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.047602 (rank : 53) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.047058 (rank : 54) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.043420 (rank : 55) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CN103_HUMAN
|
||||||
| NC score | 0.043185 (rank : 56) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96BY7, Q6ZRE7, Q9NW80 | Gene names | C14orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf103. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.042233 (rank : 57) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.041188 (rank : 58) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AMOL2_MOUSE
|
||||||
| NC score | 0.039943 (rank : 59) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
ICB1_MOUSE
|
||||||
| NC score | 0.038714 (rank : 60) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YX0 | Gene names | Icb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein homolog (ICB-1 protein). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.038261 (rank : 61) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CN103_MOUSE
|
||||||
| NC score | 0.037234 (rank : 62) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80XK6, Q8R1H5, Q9CWK6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf103 homolog. | |||||
|
SPAG4_HUMAN
|
||||||
| NC score | 0.034916 (rank : 63) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NPE6, O43648 | Gene names | SPAG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 4 protein (Outer dense fiber-associated protein SPAG4). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.033344 (rank : 64) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.032726 (rank : 65) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.031297 (rank : 66) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.031121 (rank : 67) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.031047 (rank : 68) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.030228 (rank : 69) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.030194 (rank : 70) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
SPAG4_MOUSE
|
||||||
| NC score | 0.030121 (rank : 71) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJF2 | Gene names | Spag4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 4 protein (Outer dense fiber-associated protein SPAG4). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.029071 (rank : 72) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
INP5E_HUMAN
|
||||||
| NC score | 0.028516 (rank : 73) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
HDAC6_MOUSE
|
||||||
| NC score | 0.027761 (rank : 74) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
ZNHI2_MOUSE
|
||||||
| NC score | 0.027092 (rank : 75) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY66, Q8C356, Q8VCQ7 | Gene names | Znhit2, ORF6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger HIT domain-containing protein 2 (Protein FON). | |||||
|
GP156_MOUSE
|
||||||
| NC score | 0.027011 (rank : 76) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.026336 (rank : 77) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.025985 (rank : 78) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.025604 (rank : 79) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.025557 (rank : 80) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.025448 (rank : 81) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.024451 (rank : 82) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
SEM6B_MOUSE
|
||||||
| NC score | 0.023096 (rank : 83) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.021377 (rank : 84) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
XKR7_MOUSE
|
||||||
| NC score | 0.021217 (rank : 85) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
GNL3_MOUSE
|
||||||
| NC score | 0.020968 (rank : 86) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.020929 (rank : 87) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.020404 (rank : 88) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
TDRD4_HUMAN
|
||||||
| NC score | 0.019819 (rank : 89) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
MTG8R_HUMAN
|
||||||
| NC score | 0.018819 (rank : 90) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.018095 (rank : 91) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.016854 (rank : 92) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LRRC7_MOUSE
|
||||||
| NC score | 0.016657 (rank : 93) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TE7 | Gene names | Lrrc7, Kiaa1365, Lap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.016311 (rank : 94) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.015435 (rank : 95) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.015348 (rank : 96) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.015230 (rank : 97) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.013192 (rank : 98) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
LRRC7_HUMAN
|
||||||
| NC score | 0.013103 (rank : 99) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.012642 (rank : 100) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.009953 (rank : 101) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ANR25_MOUSE
|
||||||
| NC score | 0.009835 (rank : 102) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
PP16A_HUMAN
|
||||||
| NC score | 0.008800 (rank : 103) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.008137 (rank : 104) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.004833 (rank : 105) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
CAD12_HUMAN
|
||||||
| NC score | 0.003798 (rank : 106) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55289 | Gene names | CDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-12 precursor (Brain-cadherin) (BR-cadherin) (N-cadherin 2) (Neural type cadherin 2). | |||||
|
ZFP13_MOUSE
|
||||||
| NC score | 0.003497 (rank : 107) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.002857 (rank : 108) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
ZN710_MOUSE
|
||||||
| NC score | 0.001855 (rank : 109) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.001618 (rank : 110) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
SRC_MOUSE
|
||||||
| NC score | 0.000629 (rank : 111) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05480 | Gene names | Src | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal proto-oncogene tyrosine-protein kinase Src (EC 2.7.10.2) (p60-Src) (c-Src) (pp60c-src). | |||||