Please be patient as the page loads
|
HEMGN_HUMAN
|
||||||
| SwissProt Accessions | Q9BXL5, Q6XAR3, Q86XY5, Q9NPC0 | Gene names | HEMGN, EDAG, NDR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemogen (Hemopoietic gene protein) (Negative differentiation regulator protein) (Erythroid differentiation-associated gene protein) (EDAG-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HEMGN_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9BXL5, Q6XAR3, Q86XY5, Q9NPC0 | Gene names | HEMGN, EDAG, NDR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemogen (Hemopoietic gene protein) (Negative differentiation regulator protein) (Erythroid differentiation-associated gene protein) (EDAG-1). | |||||
|
HEMGN_MOUSE
|
||||||
| θ value | 1.40038e-85 (rank : 2) | NC score | 0.841782 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERZ0, Q3TAE6, Q80YT2, Q8CF29 | Gene names | Hemgn, Ndr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemogen (Hemopoietic gene protein) (Negative differentiation regulator protein) (mNDR). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 3) | NC score | 0.173449 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 4) | NC score | 0.043334 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 5) | NC score | 0.087315 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 6) | NC score | 0.128006 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.047110 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.077288 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.104648 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
ICAL_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.120990 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.104741 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.060607 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.101748 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.066159 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.080526 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.027953 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
INVO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.061950 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
LNP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.069742 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.092612 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.111313 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.073323 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.101716 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CMTA1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.058788 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.043439 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.049338 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.023421 (rank : 99) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.084370 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.088189 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RAE1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.041503 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.063935 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.031269 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.067038 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.030123 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.080662 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZBTB6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.008897 (rank : 124) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K088, Q8BLM4 | Gene names | Zbtb6, Zfp482, Znf482 | |||
|
Domain Architecture |
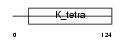
|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.036903 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
INGR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.041923 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15260 | Gene names | IFNGR1 | |||
|
Domain Architecture |
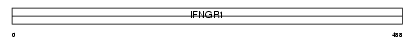
|
|||||
| Description | Interferon-gamma receptor alpha chain precursor (IFN-gamma-R1) (CD119 antigen) (CDw119). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.067199 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SCG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.062529 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.049589 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.025594 (rank : 94) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.032731 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.024726 (rank : 96) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
KIF11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.013930 (rank : 117) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |
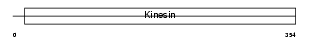
|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.080595 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PSD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.021142 (rank : 105) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLR5, Q3TE33, Q3TQF5, Q3UD59, Q3UFH9, Q80V44 | Gene names | Psd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4). | |||||
|
SH22A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.026815 (rank : 93) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.091122 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.027230 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.046350 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
GEMI5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.018086 (rank : 111) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TEQ6, Q8WWV4, Q969W4, Q9H9K3, Q9UFI5 | Gene names | GEMIN5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
JPH3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.023533 (rank : 98) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
PARPT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.020662 (rank : 107) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.056010 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.032774 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.047327 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANR25_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.022087 (rank : 102) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.012918 (rank : 118) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.012824 (rank : 120) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
ES8L2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.014098 (rank : 116) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99K30, Q91VT7 | Gene names | Eps8l2, Eps8r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
FEZ1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.024082 (rank : 97) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99689, O00679, O00728, Q6IBI7 | Gene names | FEZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
HXC10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.012043 (rank : 121) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
INP4B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.017999 (rank : 113) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15327, Q6IN59, Q6PJB4 | Gene names | INPP4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type II). | |||||
|
JPH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.025590 (rank : 95) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
KCC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.005475 (rank : 128) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08414, Q61381 | Gene names | Camk4 | |||
|
Domain Architecture |
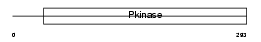
|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type IV (EC 2.7.11.17) (CAM kinase-GR) (CaMK IV). | |||||
|
KLC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.021131 (rank : 106) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
KLC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.019577 (rank : 109) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
NASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.077686 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.053967 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZFP91_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.005104 (rank : 129) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62511, Q8BPY3, Q8C2B4 | Gene names | Zfp91, Pzf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 (Zfp-91) (Zinc finger protein PZF) (Penta Zf protein). | |||||
|
1B52_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.003249 (rank : 131) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30490, Q9MY75, Q9TPT4 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |
|
|||||
| Description | HLA class I histocompatibility antigen, B-52 alpha chain precursor (MHC class I antigen B*52) (Bw-52) (B-5). | |||||
|
1B53_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.003256 (rank : 130) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30491, O78053, O78138, Q30198, Q9BD43, Q9GJF0, Q9MY42, Q9TP36, Q9TQH8 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |
|
|||||
| Description | HLA class I histocompatibility antigen, B-53 alpha chain precursor (MHC class I antigen B*53) (Bw-53). | |||||
|
AN32E_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.015824 (rank : 115) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.036235 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BUB1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.019014 (rank : 110) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.067870 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CM35H_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.021183 (rank : 104) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGN4, O95100, Q9HD97, Q9UBK4, Q9UMS9, Q9UMT0 | Gene names | CD300A, CMRF35H | |||
|
Domain Architecture |
|
|||||
| Description | CMRF35-H antigen precursor (CMRF35-H9) (CMRF-35-H9) (CD300a antigen) (Inhibitory receptor protein 60) (IRp60) (IRC1/IRC2) (NK inhibitory receptor). | |||||
|
CTGE3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.020641 (rank : 108) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX95 | Gene names | CTAGE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 3 (cTAGE-3 protein). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.011274 (rank : 122) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DEN2C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.012846 (rank : 119) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68D51, Q5TCX6, Q6P3R3 | Gene names | DENND2C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
K0460_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.038929 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
NC6IP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.028835 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
ZBTB6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.005867 (rank : 127) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.022985 (rank : 100) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
BTNL8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.005998 (rank : 126) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UX41, Q9H730 | Gene names | BTNL8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin-like protein 8 precursor. | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.022961 (rank : 101) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CEA20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.016784 (rank : 114) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UY09 | Gene names | CEACAM20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 20 precursor. | |||||
|
CO5A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.009415 (rank : 123) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
K1C10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.017999 (rank : 112) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.031830 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
UB7I1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.021809 (rank : 103) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
ZFP91_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.006622 (rank : 125) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.058566 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.060344 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.056059 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
ENL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.052030 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
ICAL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.067056 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.062880 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.060722 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K1210_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.061495 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
KI67_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.054577 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.056575 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.067007 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.066759 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.064460 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.077408 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.056547 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.067968 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.073223 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.052243 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.050523 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.071578 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.070714 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NP1L3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.050762 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051940 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.053170 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.057610 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.050798 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.056626 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RPC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051783 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
SPTA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.086119 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D9T6, Q80ZS4, Q9D5L5, Q9DAF5 | Gene names | Spata3, Tsarg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 3 (Testis spermatocyte apoptosis- related protein 1) (Testis and spermatogenesis cell-related protein 1) (mTSARG1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.055414 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TAU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.055508 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.053604 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.052029 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.071940 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.069133 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.070094 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.052545 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.090690 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.070889 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
HEMGN_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9BXL5, Q6XAR3, Q86XY5, Q9NPC0 | Gene names | HEMGN, EDAG, NDR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemogen (Hemopoietic gene protein) (Negative differentiation regulator protein) (Erythroid differentiation-associated gene protein) (EDAG-1). | |||||
|
HEMGN_MOUSE
|
||||||
| NC score | 0.841782 (rank : 2) | θ value | 1.40038e-85 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERZ0, Q3TAE6, Q80YT2, Q8CF29 | Gene names | Hemgn, Ndr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemogen (Hemopoietic gene protein) (Negative differentiation regulator protein) (mNDR). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.173449 (rank : 3) | θ value | 1.69304e-06 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.128006 (rank : 4) | θ value | 0.00228821 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.120990 (rank : 5) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.111313 (rank : 6) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.104741 (rank : 7) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.104648 (rank : 8) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.101748 (rank : 9) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.101716 (rank : 10) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.092612 (rank : 11) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.091122 (rank : 12) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.090690 (rank : 13) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.088189 (rank : 14) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.087315 (rank : 15) | θ value | 0.00134147 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
SPTA3_MOUSE
|
||||||
| NC score | 0.086119 (rank : 16) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D9T6, Q80ZS4, Q9D5L5, Q9DAF5 | Gene names | Spata3, Tsarg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 3 (Testis spermatocyte apoptosis- related protein 1) (Testis and spermatogenesis cell-related protein 1) (mTSARG1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.084370 (rank : 17) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.080662 (rank : 18) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.080595 (rank : 19) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.080526 (rank : 20) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.077686 (rank : 21) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.077408 (rank : 22) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.077288 (rank : 23) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.073323 (rank : 24) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.073223 (rank : 25) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.071940 (rank : 26) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.071578 (rank : 27) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.070889 (rank : 28) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.070714 (rank : 29) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.070094 (rank : 30) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
LNP_MOUSE
|
||||||
| NC score | 0.069742 (rank : 31) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.069133 (rank : 32) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.067968 (rank : 33) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.067870 (rank : 34) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.067199 (rank : 35) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.067056 (rank : 36) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.067038 (rank : 37) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.067007 (rank : 38) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.066759 (rank : 39) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.066159 (rank : 40) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.064460 (rank : 41) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.063935 (rank : 42) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.062880 (rank : 43) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SCG1_HUMAN
|
||||||
| NC score | 0.062529 (rank : 44) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.061950 (rank : 45) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.061495 (rank : 46) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.060722 (rank : 47) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.060607 (rank : 48) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.060344 (rank : 49) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CMTA1_HUMAN
|
||||||
| NC score | 0.058788 (rank : 50) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.058566 (rank : 51) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.057610 (rank : 52) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.056626 (rank : 53) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.056575 (rank : 54) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.056547 (rank : 55) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.056059 (rank : 56) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.056010 (rank : 57) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.055508 (rank : 58) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.055414 (rank : 59) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.054577 (rank : 60) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.053967 (rank : 61) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.053604 (rank : 62) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.053170 (rank : 63) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.052545 (rank : 64) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.052243 (rank : 65) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.052030 (rank : 66) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.052029 (rank : 67) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.051940 (rank : 68) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
RPC1_HUMAN
|
||||||
| NC score | 0.051783 (rank : 69) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.050798 (rank : 70) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NP1L3_HUMAN
|
||||||
| NC score | 0.050762 (rank : 71) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.050523 (rank : 72) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.049589 (rank : 73) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.049338 (rank : 74) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.047327 (rank : 75) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.047110 (rank : 76) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.046350 (rank : 77) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.043439 (rank : 78) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.043334 (rank : 79) | θ value | 4.1701e-05 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
INGR1_HUMAN
|
||||||
| NC score | 0.041923 (rank : 80) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15260 | Gene names | IFNGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-gamma receptor alpha chain precursor (IFN-gamma-R1) (CD119 antigen) (CDw119). | |||||
|
RAE1_MOUSE
|
||||||
| NC score | 0.041503 (rank : 81) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.038929 (rank : 82) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.036903 (rank : 83) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.036235 (rank : 84) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.032774 (rank : 85) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.032731 (rank : 86) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.031830 (rank : 87) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.031269 (rank : 88) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.030123 (rank : 89) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
NC6IP_MOUSE
|
||||||
| NC score | 0.028835 (rank : 90) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.027953 (rank : 91) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.027230 (rank : 92) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
SH22A_MOUSE
|
||||||
| NC score | 0.026815 (rank : 93) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.025594 (rank : 94) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
JPH3_HUMAN
|
||||||
| NC score | 0.025590 (rank : 95) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.024726 (rank : 96) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
FEZ1_HUMAN
|
||||||
| NC score | 0.024082 (rank : 97) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99689, O00679, O00728, Q6IBI7 | Gene names | FEZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
JPH3_MOUSE
|
||||||
| NC score | 0.023533 (rank : 98) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.023421 (rank : 99) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.022985 (rank : 100) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.022961 (rank : 101) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
ANR25_MOUSE
|
||||||
| NC score | 0.022087 (rank : 102) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
UB7I1_HUMAN
|
||||||
| NC score | 0.021809 (rank : 103) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
CM35H_HUMAN
|
||||||
| NC score | 0.021183 (rank : 104) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGN4, O95100, Q9HD97, Q9UBK4, Q9UMS9, Q9UMT0 | Gene names | CD300A, CMRF35H | |||
|
Domain Architecture |

|
|||||
| Description | CMRF35-H antigen precursor (CMRF35-H9) (CMRF-35-H9) (CD300a antigen) (Inhibitory receptor protein 60) (IRp60) (IRC1/IRC2) (NK inhibitory receptor). | |||||
|
PSD4_MOUSE
|
||||||
| NC score | 0.021142 (rank : 105) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLR5, Q3TE33, Q3TQF5, Q3UD59, Q3UFH9, Q80V44 | Gene names | Psd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4). | |||||
|
KLC3_HUMAN
|
||||||
| NC score | 0.021131 (rank : 106) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
PARPT_HUMAN
|
||||||
| NC score | 0.020662 (rank : 107) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
CTGE3_HUMAN
|
||||||
| NC score | 0.020641 (rank : 108) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX95 | Gene names | CTAGE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 3 (cTAGE-3 protein). | |||||
|
KLC3_MOUSE
|
||||||
| NC score | 0.019577 (rank : 109) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
BUB1B_MOUSE
|
||||||
| NC score | 0.019014 (rank : 110) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
GEMI5_HUMAN
|
||||||
| NC score | 0.018086 (rank : 111) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TEQ6, Q8WWV4, Q969W4, Q9H9K3, Q9UFI5 | Gene names | GEMIN5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
K1C10_MOUSE
|
||||||
| NC score | 0.017999 (rank : 112) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
INP4B_HUMAN
|
||||||
| NC score | 0.017999 (rank : 113) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15327, Q6IN59, Q6PJB4 | Gene names | INPP4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type II). | |||||
|
CEA20_HUMAN
|
||||||
| NC score | 0.016784 (rank : 114) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UY09 | Gene names | CEACAM20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 20 precursor. | |||||
|
AN32E_HUMAN
|
||||||
| NC score | 0.015824 (rank : 115) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
ES8L2_MOUSE
|
||||||
| NC score | 0.014098 (rank : 116) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99K30, Q91VT7 | Gene names | Eps8l2, Eps8r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
KIF11_HUMAN
|
||||||
| NC score | 0.013930 (rank : 117) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.012918 (rank : 118) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DEN2C_HUMAN
|
||||||
| NC score | 0.012846 (rank : 119) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68D51, Q5TCX6, Q6P3R3 | Gene names | DENND2C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2C. | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.012824 (rank : 120) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
HXC10_HUMAN
|
||||||
| NC score | 0.012043 (rank : 121) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.011274 (rank : 122) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
CO5A1_HUMAN
|
||||||
| NC score | 0.009415 (rank : 123) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
ZBTB6_MOUSE
|
||||||
| NC score | 0.008897 (rank : 124) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K088, Q8BLM4 | Gene names | Zbtb6, Zfp482, Znf482 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482). | |||||
|
ZFP91_HUMAN
|
||||||
| NC score | 0.006622 (rank : 125) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
BTNL8_HUMAN
|
||||||
| NC score | 0.005998 (rank : 126) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UX41, Q9H730 | Gene names | BTNL8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin-like protein 8 precursor. | |||||
|
ZBTB6_HUMAN
|
||||||
| NC score | 0.005867 (rank : 127) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||
|
KCC4_MOUSE
|
||||||
| NC score | 0.005475 (rank : 128) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08414, Q61381 | Gene names | Camk4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type IV (EC 2.7.11.17) (CAM kinase-GR) (CaMK IV). | |||||
|
ZFP91_MOUSE
|
||||||
| NC score | 0.005104 (rank : 129) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62511, Q8BPY3, Q8C2B4 | Gene names | Zfp91, Pzf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 (Zfp-91) (Zinc finger protein PZF) (Penta Zf protein). | |||||
|
1B53_HUMAN
|
||||||
| NC score | 0.003256 (rank : 130) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30491, O78053, O78138, Q30198, Q9BD43, Q9GJF0, Q9MY42, Q9TP36, Q9TQH8 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |

|
|||||
| Description | HLA class I histocompatibility antigen, B-53 alpha chain precursor (MHC class I antigen B*53) (Bw-53). | |||||
|
1B52_HUMAN
|
||||||
| NC score | 0.003249 (rank : 131) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30490, Q9MY75, Q9TPT4 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |

|
|||||
| Description | HLA class I histocompatibility antigen, B-52 alpha chain precursor (MHC class I antigen B*52) (Bw-52) (B-5). | |||||