Please be patient as the page loads
|
PSD4_MOUSE
|
||||||
| SwissProt Accessions | Q8BLR5, Q3TE33, Q3TQF5, Q3UD59, Q3UFH9, Q80V44 | Gene names | Psd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PSD4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.970280 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
PSD4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BLR5, Q3TE33, Q3TQF5, Q3UD59, Q3UFH9, Q80V44 | Gene names | Psd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4). | |||||
|
PSD3_HUMAN
|
||||||
| θ value | 1.8752e-114 (rank : 3) | NC score | 0.956069 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 4) | NC score | 0.727903 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 5) | NC score | 0.726804 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH3_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 6) | NC score | 0.722236 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
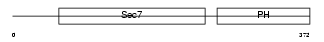
CYH1_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 7) | NC score | 0.724219 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 8) | NC score | 0.721127 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH3_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 9) | NC score | 0.725632 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH4_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 10) | NC score | 0.720450 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
CYH4_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 11) | NC score | 0.719851 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
IQEC1_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 12) | NC score | 0.701152 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6DN90, O94863, Q96D85 | Gene names | IQSEC1, ARFGEP100, BRAG2, KIAA0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1 (ADP-ribosylation factors guanine nucleotide-exchange protein 100) (ADP-ribosylation factors guanine nucleotide-exchange protein 2) (Brefeldin-resistant Arf-GEF 2 protein). | |||||
|
IQEC1_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 13) | NC score | 0.701455 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R0S2, Q3TZC3, Q5DU15 | Gene names | Iqsec1, Kiaa0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1. | |||||
|
GBF1_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 14) | NC score | 0.720633 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
BIG1_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 15) | NC score | 0.703223 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6D6, Q9NV46, Q9UFV2, Q9UNL0 | Gene names | ARFGEF1, ARFGEP1, BIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (Brefeldin A-inhibited GEP 1) (p200 ARF-GEP1) (p200 ARF guanine nucleotide exchange factor). | |||||
|
IQEC3_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 16) | NC score | 0.705578 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPP2, Q8TB43 | Gene names | IQSEC3, KIAA1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 17) | NC score | 0.697697 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 18) | NC score | 0.657599 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 19) | NC score | 0.656246 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
BIG2_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 20) | NC score | 0.691203 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 21) | NC score | 0.152628 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 22) | NC score | 0.131273 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 23) | NC score | 0.130194 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 24) | NC score | 0.145109 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 25) | NC score | 0.137120 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
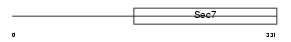
FBX8_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 26) | NC score | 0.614144 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NRD0, Q6UWN4, Q9NRP5, Q9UKC4 | Gene names | FBXO8, FBS, FBX8 | |||
|
Domain Architecture |
|
|||||
| Description | F-box only protein 8 (F-box/SEC7 protein FBS). | |||||
|
FBX8_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 27) | NC score | 0.615475 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QZN3 | Gene names | Fbxo8, Fbx8 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 8. | |||||
|
TIAM1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 28) | NC score | 0.139562 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.190369 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.197075 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.023608 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
PUNC_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.012666 (rank : 97) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVU1, O95215 | Gene names | PUNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.236210 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.186479 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
JPH4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.018914 (rank : 90) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
PLEK_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.189861 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.011081 (rank : 105) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
CA156_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.028579 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZ09, Q810I8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf156 homolog. | |||||
|
DLGP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.014835 (rank : 94) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D415, Q52KF6, Q5DTK5, Q6P6N4, Q6XBF4, Q8BZL7, Q8BZQ1, Q8C0G0 | Gene names | Dlgap1, Kiaa4162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD-95/SAP90- binding protein 1). | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.020186 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
MORC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.015181 (rank : 93) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVL5 | Gene names | Morc1, Morc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 1 (Protein microrchidia). | |||||
|
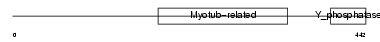
MTMR3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.012421 (rank : 99) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |
|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
PUNC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.010636 (rank : 106) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
WEE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.003064 (rank : 116) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30291 | Gene names | WEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase) (WEE1hu). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.011482 (rank : 103) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.012521 (rank : 98) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.134708 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
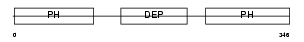
PLEK2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.126504 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin-2. | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.029232 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
GPI8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.018839 (rank : 91) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXY9, Q8BH63 | Gene names | Pigk | |||
|
Domain Architecture |
|
|||||
| Description | GPI-anchor transamidase precursor (EC 3.-.-.-) (GPI transamidase) (Phosphatidylinositol-glycan biosynthesis class K protein) (PIG-K). | |||||
|
HEMGN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.021142 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXL5, Q6XAR3, Q86XY5, Q9NPC0 | Gene names | HEMGN, EDAG, NDR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemogen (Hemopoietic gene protein) (Negative differentiation regulator protein) (Erythroid differentiation-associated gene protein) (EDAG-1). | |||||
|
BCL6B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | -0.002127 (rank : 125) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N143, Q6PCB4 | Gene names | BCL6B, BAZF, ZNF62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 6 member B protein (Bcl6-associated zinc finger protein) (Zinc finger protein 62). | |||||
|
DGKH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.037617 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.015479 (rank : 92) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PARP9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.012823 (rank : 96) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IXQ6, Q8TCP3, Q9BZL8, Q9BZL9 | Gene names | PARP9, BAL | |||
|
Domain Architecture |
|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein). | |||||
|
ZN688_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | -0.001567 (rank : 123) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
AHTF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.012398 (rank : 100) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
FER_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | -0.001950 (rank : 124) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16591 | Gene names | FER, TYK3 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FER (EC 2.7.10.2) (p94-FER) (c- FER). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.011481 (rank : 104) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.004704 (rank : 113) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MAPK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | -0.000756 (rank : 121) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49137, Q8IYD6 | Gene names | MAPKAPK2 | |||
|
Domain Architecture |
|
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
MAPK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | -0.000746 (rank : 120) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49138, Q6P561 | Gene names | Mapkapk2, Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
PLEK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.124819 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin-2. | |||||
|
E41LB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.004159 (rank : 115) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
GLI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | -0.001484 (rank : 122) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.001949 (rank : 118) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
NT5D1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.012309 (rank : 101) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TFE4, Q6XYD5 | Gene names | NT5DC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-nucleotidase domain-containing protein 1. | |||||
|
PER1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.006397 (rank : 109) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PO3F4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.005134 (rank : 111) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62515, P25216 | Gene names | Pou3f4, Brn-4, Brn4, Otf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 3, transcription factor 4 (Brain-specific homeobox/POU domain protein 4) (Brain-4) (Brn-4). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.012902 (rank : 95) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
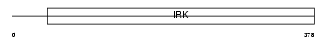
IRK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.002651 (rank : 117) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48549, Q8TBI0 | Gene names | KCNJ3, GIRK1 | |||
|
Domain Architecture |
|
|||||
| Description | G protein-activated inward rectifier potassium channel 1 (GIRK1) (Potassium channel, inwardly rectifying subfamily J member 3) (Inward rectifier K(+) channel Kir3.1). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.006699 (rank : 107) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.005653 (rank : 110) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
PCSK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.004221 (rank : 114) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29121, Q62094 | Gene names | Pcsk4, Nec-3, Nec3 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 4 precursor (EC 3.4.21.-) (PC4) (Neuroendocrine convertase 3) (NEC 3) (Prohormone convertase 3) (KEX2-like endoprotease 3). | |||||
|
PO3F4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.004718 (rank : 112) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49335, Q5H9G9, Q99410 | Gene names | POU3F4, BRN4, OTF9 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 4 (Brain-specific homeobox/POU domain protein 4) (Brain-4) (Brn-4). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.006413 (rank : 108) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.011672 (rank : 102) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.001061 (rank : 119) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
3BP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.102625 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.106412 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
ACTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.057275 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.057486 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.055542 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.055549 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.056698 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.056305 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.057029 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.057069 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.103394 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
ANLN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.084580 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
CENB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.060901 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.055868 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.057458 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CLMN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.057852 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.111930 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.112929 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
DAPP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.101546 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |
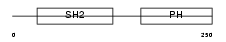
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
DAPP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.106488 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |
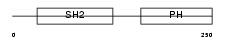
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.061900 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.059819 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.051524 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.055317 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.051133 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.050477 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.146341 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.142621 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.115399 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
PKHA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.131289 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.118447 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.128252 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.050180 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |
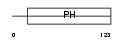
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PKHB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.054655 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96CS7, Q86W37, Q9BV75, Q9NWK1 | Gene names | PLEKHB2, EVT2 | |||
|
Domain Architecture |
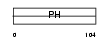
|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
PKHB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053973 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZC7, Q3UDH6, Q8C4I4, Q8CA27 | Gene names | Plekhb2, Evt2 | |||
|
Domain Architecture |
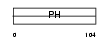
|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
RASA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.072352 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.072691 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.078072 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SMOO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.054654 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.058505 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.060313 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.054896 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.080656 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.079714 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.071172 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.072075 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.063736 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
PSD4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BLR5, Q3TE33, Q3TQF5, Q3UD59, Q3UFH9, Q80V44 | Gene names | Psd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4). | |||||
|
PSD4_HUMAN
|
||||||
| NC score | 0.970280 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
PSD3_HUMAN
|
||||||
| NC score | 0.956069 (rank : 3) | θ value | 1.8752e-114 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.727903 (rank : 4) | θ value | 1.05554e-24 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.726804 (rank : 5) | θ value | 5.23862e-24 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH3_HUMAN
|
||||||
| NC score | 0.725632 (rank : 6) | θ value | 9.87957e-23 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH1_MOUSE
|
||||||
| NC score | 0.724219 (rank : 7) | θ value | 4.43474e-23 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.722236 (rank : 8) | θ value | 8.93572e-24 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.721127 (rank : 9) | θ value | 5.79196e-23 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
GBF1_HUMAN
|
||||||
| NC score | 0.720633 (rank : 10) | θ value | 3.28887e-18 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
CYH4_MOUSE
|
||||||
| NC score | 0.720450 (rank : 11) | θ value | 2.20094e-22 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
CYH4_HUMAN
|
||||||
| NC score | 0.719851 (rank : 12) | θ value | 1.42661e-21 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
IQEC3_HUMAN
|
||||||
| NC score | 0.705578 (rank : 13) | θ value | 2.7842e-17 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPP2, Q8TB43 | Gene names | IQSEC3, KIAA1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
BIG1_HUMAN
|
||||||
| NC score | 0.703223 (rank : 14) | θ value | 9.56915e-18 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6D6, Q9NV46, Q9UFV2, Q9UNL0 | Gene names | ARFGEF1, ARFGEP1, BIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (Brefeldin A-inhibited GEP 1) (p200 ARF-GEP1) (p200 ARF guanine nucleotide exchange factor). | |||||
|
IQEC1_MOUSE
|
||||||
| NC score | 0.701455 (rank : 15) | θ value | 1.47631e-18 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R0S2, Q3TZC3, Q5DU15 | Gene names | Iqsec1, Kiaa0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1. | |||||
|
IQEC1_HUMAN
|
||||||
| NC score | 0.701152 (rank : 16) | θ value | 8.65492e-19 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6DN90, O94863, Q96D85 | Gene names | IQSEC1, ARFGEP100, BRAG2, KIAA0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1 (ADP-ribosylation factors guanine nucleotide-exchange protein 100) (ADP-ribosylation factors guanine nucleotide-exchange protein 2) (Brefeldin-resistant Arf-GEF 2 protein). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.697697 (rank : 17) | θ value | 2.7842e-17 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
BIG2_HUMAN
|
||||||
| NC score | 0.691203 (rank : 18) | θ value | 2.60593e-15 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.657599 (rank : 19) | θ value | 6.85773e-16 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.656246 (rank : 20) | θ value | 1.52774e-15 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
FBX8_MOUSE
|
||||||
| NC score | 0.615475 (rank : 21) | θ value | 1.09739e-05 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QZN3 | Gene names | Fbxo8, Fbx8 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 8. | |||||
|
FBX8_HUMAN
|
||||||
| NC score | 0.614144 (rank : 22) | θ value | 1.09739e-05 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NRD0, Q6UWN4, Q9NRP5, Q9UKC4 | Gene names | FBXO8, FBS, FBX8 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 8 (F-box/SEC7 protein FBS). | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.236210 (rank : 23) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.197075 (rank : 24) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.190369 (rank : 25) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.189861 (rank : 26) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.186479 (rank : 27) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.152628 (rank : 28) | θ value | 6.64225e-11 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.146341 (rank : 29) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.145109 (rank : 30) | θ value | 1.69304e-06 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.142621 (rank : 31) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
TIAM1_MOUSE
|
||||||
| NC score | 0.139562 (rank : 32) | θ value | 1.43324e-05 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.137120 (rank : 33) | θ value | 2.88788e-06 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.134708 (rank : 34) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PKHA2_MOUSE
|
||||||
| NC score | 0.131289 (rank : 35) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.131273 (rank : 36) | θ value | 2.79066e-09 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.130194 (rank : 37) | θ value | 4.76016e-09 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.128252 (rank : 38) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PLEK2_MOUSE
|
||||||
| NC score | 0.126504 (rank : 39) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK2_HUMAN
|
||||||
| NC score | 0.124819 (rank : 40) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.118447 (rank : 41) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA2_HUMAN
|
||||||
| NC score | 0.115399 (rank : 42) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.112929 (rank : 43) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.111930 (rank : 44) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
DAPP1_MOUSE
|
||||||
| NC score | 0.106488 (rank : 45) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.106412 (rank : 46) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.103394 (rank : 47) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.102625 (rank : 48) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
DAPP1_HUMAN
|
||||||
| NC score | 0.101546 (rank : 49) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
ANLN_MOUSE
|
||||||
| NC score | 0.084580 (rank : 50) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.080656 (rank : 51) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.079714 (rank : 52) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.078072 (rank : 53) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.072691 (rank : 54) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RASA1_HUMAN
|
||||||
| NC score | 0.072352 (rank : 55) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.072075 (rank : 56) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.071172 (rank : 57) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.063736 (rank : 58) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.061900 (rank : 59) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.060901 (rank : 60) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.060313 (rank : 61) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.059819 (rank : 62) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.058505 (rank : 63) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.057852 (rank : 64) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
ACTN1_MOUSE
|
||||||
| NC score | 0.057486 (rank : 65) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.057458 (rank : 66) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
ACTN1_HUMAN
|
||||||
| NC score | 0.057275 (rank : 67) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN4_MOUSE
|
||||||
| NC score | 0.057069 (rank : 68) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN4_HUMAN
|
||||||
| NC score | 0.057029 (rank : 69) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN3_HUMAN
|
||||||
| NC score | 0.056698 (rank : 70) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| NC score | 0.056305 (rank : 71) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.055868 (rank : 72) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
ACTN2_MOUSE
|
||||||
| NC score | 0.055549 (rank : 73) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_HUMAN
|
||||||
| NC score | 0.055542 (rank : 74) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.055317 (rank : 75) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.054896 (rank : 76) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
PKHB2_HUMAN
|
||||||
| NC score | 0.054655 (rank : 77) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96CS7, Q86W37, Q9BV75, Q9NWK1 | Gene names | PLEKHB2, EVT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.054654 (rank : 78) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
PKHB2_MOUSE
|
||||||
| NC score | 0.053973 (rank : 79) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZC7, Q3UDH6, Q8C4I4, Q8CA27 | Gene names | Plekhb2, Evt2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.051524 (rank : 80) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.051133 (rank : 81) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.050477 (rank : 82) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
PKHB1_HUMAN
|
||||||
| NC score | 0.050180 (rank : 83) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
DGKH_HUMAN
|
||||||
| NC score | 0.037617 (rank : 84) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.029232 (rank : 85) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
CA156_MOUSE
|
||||||
| NC score | 0.028579 (rank : 86) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZ09, Q810I8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf156 homolog. | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.023608 (rank : 87) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
HEMGN_HUMAN
|
||||||
| NC score | 0.021142 (rank : 88) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXL5, Q6XAR3, Q86XY5, Q9NPC0 | Gene names | HEMGN, EDAG, NDR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemogen (Hemopoietic gene protein) (Negative differentiation regulator protein) (Erythroid differentiation-associated gene protein) (EDAG-1). | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.020186 (rank : 89) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
JPH4_HUMAN
|
||||||
| NC score | 0.018914 (rank : 90) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
GPI8_MOUSE
|
||||||
| NC score | 0.018839 (rank : 91) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXY9, Q8BH63 | Gene names | Pigk | |||
|
Domain Architecture |

|
|||||
| Description | GPI-anchor transamidase precursor (EC 3.-.-.-) (GPI transamidase) (Phosphatidylinositol-glycan biosynthesis class K protein) (PIG-K). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.015479 (rank : 92) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
MORC1_MOUSE
|
||||||
| NC score | 0.015181 (rank : 93) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVL5 | Gene names | Morc1, Morc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 1 (Protein microrchidia). | |||||
|
DLGP1_MOUSE
|
||||||
| NC score | 0.014835 (rank : 94) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D415, Q52KF6, Q5DTK5, Q6P6N4, Q6XBF4, Q8BZL7, Q8BZQ1, Q8C0G0 | Gene names | Dlgap1, Kiaa4162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD-95/SAP90- binding protein 1). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.012902 (rank : 95) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
PARP9_HUMAN
|
||||||
| NC score | 0.012823 (rank : 96) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IXQ6, Q8TCP3, Q9BZL8, Q9BZL9 | Gene names | PARP9, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein). | |||||
|
PUNC_HUMAN
|
||||||
| NC score | 0.012666 (rank : 97) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVU1, O95215 | Gene names | PUNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.012521 (rank : 98) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
MTMR3_MOUSE
|
||||||
| NC score | 0.012421 (rank : 99) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
AHTF1_MOUSE
|
||||||
| NC score | 0.012398 (rank : 100) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
NT5D1_HUMAN
|
||||||
| NC score | 0.012309 (rank : 101) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TFE4, Q6XYD5 | Gene names | NT5DC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-nucleotidase domain-containing protein 1. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.011672 (rank : 102) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.011482 (rank : 103) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.011481 (rank : 104) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.011081 (rank : 105) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
PUNC_MOUSE
|
||||||
| NC score | 0.010636 (rank : 106) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.006699 (rank : 107) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.006413 (rank : 108) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.006397 (rank : 109) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.005653 (rank : 110) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
PO3F4_MOUSE
|
||||||
| NC score | 0.005134 (rank : 111) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62515, P25216 | Gene names | Pou3f4, Brn-4, Brn4, Otf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 3, transcription factor 4 (Brain-specific homeobox/POU domain protein 4) (Brain-4) (Brn-4). | |||||
|
PO3F4_HUMAN
|
||||||
| NC score | 0.004718 (rank : 112) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49335, Q5H9G9, Q99410 | Gene names | POU3F4, BRN4, OTF9 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 4 (Brain-specific homeobox/POU domain protein 4) (Brain-4) (Brn-4). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.004704 (rank : 113) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PCSK4_MOUSE
|
||||||
| NC score | 0.004221 (rank : 114) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29121, Q62094 | Gene names | Pcsk4, Nec-3, Nec3 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 4 precursor (EC 3.4.21.-) (PC4) (Neuroendocrine convertase 3) (NEC 3) (Prohormone convertase 3) (KEX2-like endoprotease 3). | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.004159 (rank : 115) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
WEE1_HUMAN
|
||||||
| NC score | 0.003064 (rank : 116) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30291 | Gene names | WEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase) (WEE1hu). | |||||
|
IRK3_HUMAN
|
||||||
| NC score | 0.002651 (rank : 117) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48549, Q8TBI0 | Gene names | KCNJ3, GIRK1 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 1 (GIRK1) (Potassium channel, inwardly rectifying subfamily J member 3) (Inward rectifier K(+) channel Kir3.1). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.001949 (rank : 118) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.001061 (rank : 119) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MAPK2_MOUSE
|
||||||
| NC score | -0.000746 (rank : 120) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49138, Q6P561 | Gene names | Mapkapk2, Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
MAPK2_HUMAN
|
||||||
| NC score | -0.000756 (rank : 121) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49137, Q8IYD6 | Gene names | MAPKAPK2 | |||
|
Domain Architecture |

|
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
GLI1_MOUSE
|
||||||
| NC score | -0.001484 (rank : 122) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
ZN688_HUMAN
|
||||||
| NC score | -0.001567 (rank : 123) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
FER_HUMAN
|
||||||
| NC score | -0.001950 (rank : 124) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16591 | Gene names | FER, TYK3 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FER (EC 2.7.10.2) (p94-FER) (c- FER). | |||||
|
BCL6B_HUMAN
|
||||||
| NC score | -0.002127 (rank : 125) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N143, Q6PCB4 | Gene names | BCL6B, BAZF, ZNF62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 6 member B protein (Bcl6-associated zinc finger protein) (Zinc finger protein 62). | |||||