Please be patient as the page loads
|
SMYD4_HUMAN
|
||||||
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMYD4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.974033 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD2_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 3) | NC score | 0.479555 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
SMYD3_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 4) | NC score | 0.485534 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD2_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 5) | NC score | 0.473494 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
SMYD3_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 6) | NC score | 0.479963 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
TTC4_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 7) | NC score | 0.400063 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |
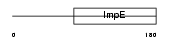
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
TTC4_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 8) | NC score | 0.403093 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |
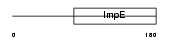
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
SMYD1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 9) | NC score | 0.410174 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
SMYD5_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 10) | NC score | 0.370001 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TYX3, Q3TRB2, Q91YL6 | Gene names | Smyd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 5 (Protein NN8-4AG). | |||||
|
SMYD1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 11) | NC score | 0.409421 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
SMYD5_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 12) | NC score | 0.361198 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6GMV2, Q13558 | Gene names | SMYD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 5 (Protein NN8-4AG). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 13) | NC score | 0.221416 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
Z3H7B_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 14) | NC score | 0.257039 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |
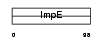
|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 15) | NC score | 0.252693 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
TTC12_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 16) | NC score | 0.250211 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |
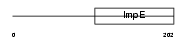
|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
UN45B_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 17) | NC score | 0.255375 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
UN45B_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 18) | NC score | 0.257991 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 19) | NC score | 0.040723 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 20) | NC score | 0.226570 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
SGTB_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 21) | NC score | 0.228827 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |
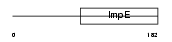
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 22) | NC score | 0.232403 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 23) | NC score | 0.226981 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
TTC1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 24) | NC score | 0.253605 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
OM34_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 25) | NC score | 0.241986 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |
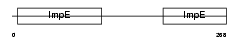
|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
ZMY15_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 26) | NC score | 0.141107 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 27) | NC score | 0.162313 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
SETD4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 28) | NC score | 0.142645 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58467 | Gene names | Setd4, ORF21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET domain-containing protein 4. | |||||
|
TTC12_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 29) | NC score | 0.231513 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 30) | NC score | 0.159937 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.228131 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
OM34_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.233630 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 33) | NC score | 0.035740 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
SETD4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.125701 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVD3, Q9BU46 | Gene names | SETD4, C21orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET domain-containing protein 4. | |||||
|
PDCD2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.117496 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
MTG8R_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.111968 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
MTG8R_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.112145 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.044856 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.031352 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.018650 (rank : 105) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
SGTA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.191281 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |
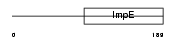
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
MTG8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.098543 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
MTG8_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.099193 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
ANKY1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.047180 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.015122 (rank : 107) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
PDCD2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.117282 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.010961 (rank : 109) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.029479 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
NEK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.001413 (rank : 120) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 908 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51955, Q53FD6, Q5VXZ1, Q6NZX8, Q7Z634, Q86XH2, Q96QN9 | Gene names | NEK2, NLK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek2 (EC 2.7.11.1) (NimA-related protein kinase 2) (NimA-like protein kinase 1) (HSPK 21). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.193224 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.015233 (rank : 106) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
K1688_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.035545 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
TTC13_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.086688 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
ZN683_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.003042 (rank : 119) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZ20, Q5T141, Q5T146, Q5T147, Q5T149, Q8NEN4 | Gene names | ZNF683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 683. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.010593 (rank : 110) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.028258 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
RFC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.023390 (rank : 102) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35250, P32846, Q9BU93 | Gene names | RFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.023423 (rank : 101) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUK4 | Gene names | Rfc2 | |||
|
Domain Architecture |
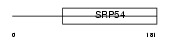
|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.028326 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ZFP60_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.000912 (rank : 123) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16374, Q61135 | Gene names | Zfp60, Mfg3 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 60 (Zfp-60) (Zinc finger protein Mfg-3). | |||||
|
CH076_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.021454 (rank : 103) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96K31, Q53HC1 | Gene names | C8orf76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C8orf76. | |||||
|
CP11A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.009590 (rank : 114) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ82 | Gene names | Cyp11a1, Cyp11a | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11A1, mitochondrial precursor (EC 1.14.15.6) (CYPXIA1) (P450(scc)) (Cholesterol side-chain cleavage enzyme) (Cholesterol desmolase). | |||||
|
KLC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.024411 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
NEK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.000366 (rank : 125) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35942, O35959 | Gene names | Nek2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek2 (EC 2.7.11.1) (NimA-related protein kinase 2). | |||||
|
ZN623_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.000583 (rank : 124) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75123 | Gene names | ZNF623, KIAA0628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 623. | |||||
|
ZXDB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.005234 (rank : 116) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98169, Q9UBB3 | Gene names | ZXDB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDB. | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.014131 (rank : 108) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CJ093_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.023856 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SR76, Q86ST2, Q8N777 | Gene names | C10orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf93. | |||||
|
FKBP8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.103319 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.008464 (rank : 115) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.019045 (rank : 104) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.009937 (rank : 113) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
SETD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.030505 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.057325 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
ZXDA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.004993 (rank : 117) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98168, Q9UJP7 | Gene names | ZXDA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDA. | |||||
|
LMNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.010051 (rank : 112) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 653 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02545, P02546, Q969I8, Q96JA2 | Gene names | LMNA, LMN1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C (70 kDa lamin) (NY-REN-32 antigen). | |||||
|
LMNA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.010174 (rank : 111) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48678, P11516, P97859, Q3TIH0, Q3TTS8, Q3UCA0, Q3UCJ8, Q91WF2, Q9DC21 | Gene names | Lmna, Lmn1 | |||
|
Domain Architecture |
|
|||||
| Description | Lamin-A/C. | |||||
|
SETD8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.027903 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
ZN509_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.003404 (rank : 118) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN710_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.001186 (rank : 122) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
ZN710_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.001209 (rank : 121) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.063772 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.060871 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.060326 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
AIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.055255 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.061670 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DEAF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.061104 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.065590 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.066664 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.124982 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
DYXC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.135599 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
EGLN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050760 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.052850 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
F10A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.074609 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
F10A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.076748 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
F10A4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.078925 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
F10A5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.074697 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
FKBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.091753 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |
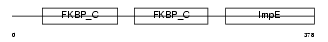
|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.089242 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.090457 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.084649 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.091554 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
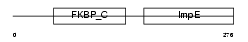
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.055797 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
PPID_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.068923 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPID_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.064144 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
PPP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.090449 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
PPP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.093355 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
SGTA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.166808 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |
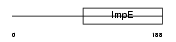
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
SGTB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.189641 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
STUB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.143431 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
STUB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.139470 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
SUGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.071536 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |
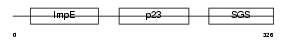
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
SUGT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.082639 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |
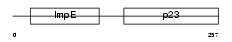
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
TOM70_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.151935 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.153993 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.057367 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
TTC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.071897 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
TTC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.073868 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
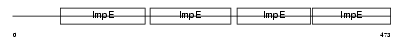
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
WDTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.074945 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.076381 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
Z3H7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.142462 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
ZMY10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.054853 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY17_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.088229 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
ZMY19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.113623 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.113623 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.974033 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD3_MOUSE
|
||||||
| NC score | 0.485534 (rank : 3) | θ value | 1.74796e-11 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_HUMAN
|
||||||
| NC score | 0.479963 (rank : 4) | θ value | 2.28291e-11 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD2_HUMAN
|
||||||
| NC score | 0.479555 (rank : 5) | θ value | 1.33837e-11 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
SMYD2_MOUSE
|
||||||
| NC score | 0.473494 (rank : 6) | θ value | 2.28291e-11 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
SMYD1_MOUSE
|
||||||
| NC score | 0.410174 (rank : 7) | θ value | 9.92553e-07 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
SMYD1_HUMAN
|
||||||
| NC score | 0.409421 (rank : 8) | θ value | 2.21117e-06 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
TTC4_MOUSE
|
||||||
| NC score | 0.403093 (rank : 9) | θ value | 7.34386e-10 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
TTC4_HUMAN
|
||||||
| NC score | 0.400063 (rank : 10) | θ value | 3.29651e-10 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
SMYD5_MOUSE
|
||||||
| NC score | 0.370001 (rank : 11) | θ value | 9.92553e-07 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TYX3, Q3TRB2, Q91YL6 | Gene names | Smyd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 5 (Protein NN8-4AG). | |||||
|
SMYD5_HUMAN
|
||||||
| NC score | 0.361198 (rank : 12) | θ value | 6.43352e-06 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6GMV2, Q13558 | Gene names | SMYD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 5 (Protein NN8-4AG). | |||||
|
UN45B_MOUSE
|
||||||
| NC score | 0.257991 (rank : 13) | θ value | 0.000786445 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
Z3H7B_HUMAN
|
||||||
| NC score | 0.257039 (rank : 14) | θ value | 9.29e-05 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
UN45B_HUMAN
|
||||||
| NC score | 0.255375 (rank : 15) | θ value | 0.000786445 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.253605 (rank : 16) | θ value | 0.0113563 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.252693 (rank : 17) | θ value | 0.000270298 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
TTC12_MOUSE
|
||||||
| NC score | 0.250211 (rank : 18) | θ value | 0.000786445 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
OM34_MOUSE
|
||||||
| NC score | 0.241986 (rank : 19) | θ value | 0.0252991 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
OM34_HUMAN
|
||||||
| NC score | 0.233630 (rank : 20) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.232403 (rank : 21) | θ value | 0.00509761 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.231513 (rank : 22) | θ value | 0.0330416 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
SGTB_HUMAN
|
||||||
| NC score | 0.228827 (rank : 23) | θ value | 0.00175202 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.228131 (rank : 24) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.226981 (rank : 25) | θ value | 0.0113563 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.226570 (rank : 26) | θ value | 0.00134147 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.221416 (rank : 27) | θ value | 9.29e-05 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.193224 (rank : 28) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
SGTA_MOUSE
|
||||||
| NC score | 0.191281 (rank : 29) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
SGTB_MOUSE
|
||||||
| NC score | 0.189641 (rank : 30) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
SGTA_HUMAN
|
||||||
| NC score | 0.166808 (rank : 31) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.162313 (rank : 32) | θ value | 0.0330416 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.159937 (rank : 33) | θ value | 0.0563607 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
TOM70_MOUSE
|
||||||
| NC score | 0.153993 (rank : 34) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.151935 (rank : 35) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.143431 (rank : 36) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
SETD4_MOUSE
|
||||||
| NC score | 0.142645 (rank : 37) | θ value | 0.0330416 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58467 | Gene names | Setd4, ORF21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET domain-containing protein 4. | |||||
|
Z3H7A_HUMAN
|
||||||
| NC score | 0.142462 (rank : 38) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
ZMY15_HUMAN
|
||||||
| NC score | 0.141107 (rank : 39) | θ value | 0.0252991 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.139470 (rank : 40) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
DYXC1_MOUSE
|
||||||
| NC score | 0.135599 (rank : 41) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
SETD4_HUMAN
|
||||||
| NC score | 0.125701 (rank : 42) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVD3, Q9BU46 | Gene names | SETD4, C21orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET domain-containing protein 4. | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.124982 (rank : 43) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
PDCD2_HUMAN
|
||||||
| NC score | 0.117496 (rank : 44) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
PDCD2_MOUSE
|
||||||
| NC score | 0.117282 (rank : 45) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
ZMY19_HUMAN
|
||||||
| NC score | 0.113623 (rank : 46) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| NC score | 0.113623 (rank : 47) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
MTG8R_MOUSE
|
||||||
| NC score | 0.112145 (rank : 48) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
MTG8R_HUMAN
|
||||||
| NC score | 0.111968 (rank : 49) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
FKBP8_MOUSE
|
||||||
| NC score | 0.103319 (rank : 50) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
MTG8_MOUSE
|
||||||
| NC score | 0.099193 (rank : 51) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
MTG8_HUMAN
|
||||||
| NC score | 0.098543 (rank : 52) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
PPP5_MOUSE
|
||||||
| NC score | 0.093355 (rank : 53) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
FKBP4_HUMAN
|
||||||
| NC score | 0.091753 (rank : 54) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.091554 (rank : 55) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
FKBP5_HUMAN
|
||||||
| NC score | 0.090457 (rank : 56) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
PPP5_HUMAN
|
||||||
| NC score | 0.090449 (rank : 57) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
FKBP4_MOUSE
|
||||||
| NC score | 0.089242 (rank : 58) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
ZMY17_MOUSE
|
||||||
| NC score | 0.088229 (rank : 59) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
TTC13_HUMAN
|
||||||
| NC score | 0.086688 (rank : 60) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
FKBP5_MOUSE
|
||||||
| NC score | 0.084649 (rank : 61) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
SUGT1_MOUSE
|
||||||
| NC score | 0.082639 (rank : 62) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.078925 (rank : 63) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
F10A1_MOUSE
|
||||||
| NC score | 0.076748 (rank : 64) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
WDTC1_MOUSE
|
||||||
| NC score | 0.076381 (rank : 65) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_HUMAN
|
||||||
| NC score | 0.074945 (rank : 66) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.074697 (rank : 67) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.074609 (rank : 68) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.073868 (rank : 69) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.071897 (rank : 70) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
SUGT1_HUMAN
|
||||||
| NC score | 0.071536 (rank : 71) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.068923 (rank : 72) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.066664 (rank : 73) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.065590 (rank : 74) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.064144 (rank : 75) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.063772 (rank : 76) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.061670 (rank : 77) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DEAF1_MOUSE
|
||||||
| NC score | 0.061104 (rank : 78) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
AIPL1_MOUSE
|
||||||
| NC score | 0.060871 (rank : 79) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIP_HUMAN
|
||||||
| NC score | 0.060326 (rank : 80) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.057367 (rank : 81) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.057325 (rank : 82) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.055797 (rank : 83) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
AIP_MOUSE
|
||||||
| NC score | 0.055255 (rank : 84) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
ZMY10_HUMAN
|
||||||
| NC score | 0.054853 (rank : 85) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.052850 (rank : 86) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
EGLN1_HUMAN
|
||||||
| NC score | 0.050760 (rank : 87) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
ANKY1_HUMAN
|
||||||
| NC score | 0.047180 (rank : 88) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.044856 (rank : 89) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.040723 (rank : 90) | θ value | 0.00134147 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.035740 (rank : 91) | θ value | 0.0961366 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.035545 (rank : 92) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.031352 (rank : 93) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SETD8_HUMAN
|
||||||
| NC score | 0.030505 (rank : 94) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.029479 (rank : 95) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.028326 (rank : 96) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.028258 (rank : 97) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
SETD8_MOUSE
|
||||||
| NC score | 0.027903 (rank : 98) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
KLC4_MOUSE
|
||||||
| NC score | 0.024411 (rank : 99) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
CJ093_HUMAN
|
||||||
| NC score | 0.023856 (rank : 100) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SR76, Q86ST2, Q8N777 | Gene names | C10orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf93. | |||||
|
RFC2_MOUSE
|
||||||
| NC score | 0.023423 (rank : 101) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUK4 | Gene names | Rfc2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC2_HUMAN
|
||||||
| NC score | 0.023390 (rank : 102) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35250, P32846, Q9BU93 | Gene names | RFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
CH076_HUMAN
|
||||||
| NC score | 0.021454 (rank : 103) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96K31, Q53HC1 | Gene names | C8orf76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C8orf76. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.019045 (rank : 104) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.018650 (rank : 105) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.015233 (rank : 106) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.015122 (rank : 107) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.014131 (rank : 108) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.010961 (rank : 109) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.010593 (rank : 110) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
LMNA_MOUSE
|
||||||
| NC score | 0.010174 (rank : 111) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48678, P11516, P97859, Q3TIH0, Q3TTS8, Q3UCA0, Q3UCJ8, Q91WF2, Q9DC21 | Gene names | Lmna, Lmn1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C. | |||||
|
LMNA_HUMAN
|
||||||
| NC score | 0.010051 (rank : 112) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 653 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02545, P02546, Q969I8, Q96JA2 | Gene names | LMNA, LMN1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C (70 kDa lamin) (NY-REN-32 antigen). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.009937 (rank : 113) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
CP11A_MOUSE
|
||||||
| NC score | 0.009590 (rank : 114) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ82 | Gene names | Cyp11a1, Cyp11a | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11A1, mitochondrial precursor (EC 1.14.15.6) (CYPXIA1) (P450(scc)) (Cholesterol side-chain cleavage enzyme) (Cholesterol desmolase). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.008464 (rank : 115) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
ZXDB_HUMAN
|
||||||
| NC score | 0.005234 (rank : 116) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98169, Q9UBB3 | Gene names | ZXDB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDB. | |||||
|
ZXDA_HUMAN
|
||||||
| NC score | 0.004993 (rank : 117) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98168, Q9UJP7 | Gene names | ZXDA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDA. | |||||
|
ZN509_HUMAN
|
||||||
| NC score | 0.003404 (rank : 118) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN683_HUMAN
|
||||||
| NC score | 0.003042 (rank : 119) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZ20, Q5T141, Q5T146, Q5T147, Q5T149, Q8NEN4 | Gene names | ZNF683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 683. | |||||
|
NEK2_HUMAN
|
||||||
| NC score | 0.001413 (rank : 120) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 908 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51955, Q53FD6, Q5VXZ1, Q6NZX8, Q7Z634, Q86XH2, Q96QN9 | Gene names | NEK2, NLK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek2 (EC 2.7.11.1) (NimA-related protein kinase 2) (NimA-like protein kinase 1) (HSPK 21). | |||||
|
ZN710_MOUSE
|
||||||
| NC score | 0.001209 (rank : 121) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
ZN710_HUMAN
|
||||||
| NC score | 0.001186 (rank : 122) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
ZFP60_MOUSE
|
||||||
| NC score | 0.000912 (rank : 123) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16374, Q61135 | Gene names | Zfp60, Mfg3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 60 (Zfp-60) (Zinc finger protein Mfg-3). | |||||
|
ZN623_HUMAN
|
||||||
| NC score | 0.000583 (rank : 124) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75123 | Gene names | ZNF623, KIAA0628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 623. | |||||
|
NEK2_MOUSE
|
||||||
| NC score | 0.000366 (rank : 125) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35942, O35959 | Gene names | Nek2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek2 (EC 2.7.11.1) (NimA-related protein kinase 2). | |||||