Please be patient as the page loads
|
TTC1_MOUSE
|
||||||
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TTC1_MOUSE
|
||||||
| θ value | 1.98038e-156 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 5.99046e-137 (rank : 2) | NC score | 0.912747 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 3) | NC score | 0.777418 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
OM34_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 4) | NC score | 0.805891 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
TOM70_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 5) | NC score | 0.772626 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 6) | NC score | 0.776716 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 7) | NC score | 0.780367 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
OM34_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 8) | NC score | 0.807309 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |
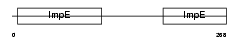
|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
UN45B_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 9) | NC score | 0.670852 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 10) | NC score | 0.580753 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 11) | NC score | 0.582866 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
UN45B_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 12) | NC score | 0.668622 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 13) | NC score | 0.671248 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 14) | NC score | 0.692967 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 15) | NC score | 0.731183 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 16) | NC score | 0.728905 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
SGTB_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 17) | NC score | 0.746564 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |
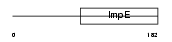
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
SGTB_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 18) | NC score | 0.745549 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
PPP5_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 19) | NC score | 0.444039 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
SGTA_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 20) | NC score | 0.711904 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |
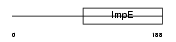
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
SGTA_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 21) | NC score | 0.714025 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |
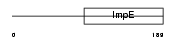
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
PPP5_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 22) | NC score | 0.445137 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
F10A4_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 23) | NC score | 0.585320 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
TTC12_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 24) | NC score | 0.711221 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
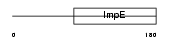
TTC4_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 25) | NC score | 0.665979 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
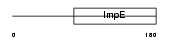
TTC4_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 26) | NC score | 0.647373 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
F10A1_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 27) | NC score | 0.552974 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 28) | NC score | 0.422422 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
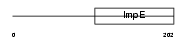
TTC12_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 29) | NC score | 0.702216 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 30) | NC score | 0.430221 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
FKBP5_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 31) | NC score | 0.517356 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
F10A5_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 32) | NC score | 0.552153 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
WDTC1_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 33) | NC score | 0.384985 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 34) | NC score | 0.383112 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
FKBP5_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 35) | NC score | 0.496683 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 36) | NC score | 0.611350 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
F10A1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 37) | NC score | 0.579938 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
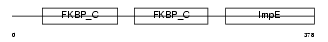
FKBP4_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 38) | NC score | 0.500812 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP8_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 39) | NC score | 0.536280 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |
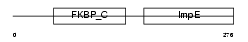
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
FKBP4_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 40) | NC score | 0.492377 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 41) | NC score | 0.520028 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
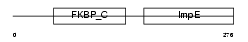
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
STUB1_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 42) | NC score | 0.698082 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
DYXC1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 43) | NC score | 0.628479 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
STUB1_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 44) | NC score | 0.688789 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
PPID_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 45) | NC score | 0.337779 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 46) | NC score | 0.417136 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 47) | NC score | 0.372558 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
AIPL1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 48) | NC score | 0.403813 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
PPID_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 49) | NC score | 0.349490 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
AIP_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 50) | NC score | 0.386171 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
Z3H7B_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 51) | NC score | 0.494460 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |
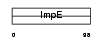
|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 52) | NC score | 0.023894 (rank : 176) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 53) | NC score | 0.237481 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
AIP_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 54) | NC score | 0.372597 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 55) | NC score | 0.325613 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 56) | NC score | 0.055972 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 57) | NC score | 0.253605 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 58) | NC score | 0.246331 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
BIN3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 59) | NC score | 0.096714 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JI08 | Gene names | Bin3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bridging integrator 3. | |||||
|
TTC3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 60) | NC score | 0.321874 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
TTC8_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 61) | NC score | 0.128100 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |
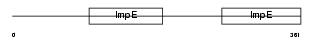
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 62) | NC score | 0.063552 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 63) | NC score | 0.068589 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 64) | NC score | 0.067276 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 65) | NC score | 0.055963 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
TTC13_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 66) | NC score | 0.365463 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
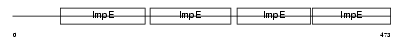
TTC6_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 67) | NC score | 0.451145 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 68) | NC score | 0.041566 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
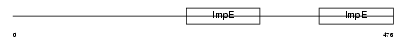
TTC8_MOUSE
|
||||||
| θ value | 0.163984 (rank : 69) | NC score | 0.114220 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
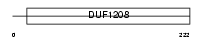
BIN3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 70) | NC score | 0.094305 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQY0, Q9BVG2, Q9NVY9 | Gene names | BIN3 | |||
|
Domain Architecture |
|
|||||
| Description | Bridging integrator 3. | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 71) | NC score | 0.059502 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
IFT88_MOUSE
|
||||||
| θ value | 0.21417 (rank : 72) | NC score | 0.202195 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 73) | NC score | 0.047504 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
K0372_HUMAN
|
||||||
| θ value | 0.279714 (rank : 74) | NC score | 0.269484 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
KIF3A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 75) | NC score | 0.042026 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 76) | NC score | 0.061694 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.365318 (rank : 77) | NC score | 0.033867 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NUF2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 78) | NC score | 0.047837 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 0.365318 (rank : 79) | NC score | 0.022626 (rank : 180) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
Z3H7A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 80) | NC score | 0.392369 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
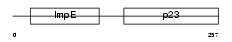
SUGT1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 81) | NC score | 0.376183 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
INCE_HUMAN
|
||||||
| θ value | 0.62314 (rank : 82) | NC score | 0.053670 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 83) | NC score | 0.043010 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 84) | NC score | 0.054044 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 85) | NC score | 0.054240 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 86) | NC score | 0.037362 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 87) | NC score | 0.061019 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.044665 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
IFT88_HUMAN
|
||||||
| θ value | 1.06291 (rank : 89) | NC score | 0.209167 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 90) | NC score | 0.059052 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 91) | NC score | 0.024638 (rank : 175) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
STAP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 92) | NC score | 0.035549 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULZ2 | Gene names | STAP1, BRDG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1) (BCR downstream signaling protein 1) (Docking protein BRDG1). | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 93) | NC score | 0.044425 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
UBE4B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 94) | NC score | 0.045506 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 95) | NC score | 0.023171 (rank : 178) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
FA50A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | 0.044430 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WV03, Q9D866 | Gene names | Fam50a, D0HXS9928E, Xap5 | |||
|
Domain Architecture |
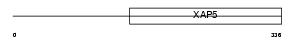
|
|||||
| Description | Protein FAM50A (XAP-5 protein). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 1.38821 (rank : 97) | NC score | 0.020355 (rank : 181) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIM44_MOUSE
|
||||||
| θ value | 1.38821 (rank : 98) | NC score | 0.034322 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35857 | Gene names | Timm44, Mimt44, Tim44 | |||
|
Domain Architecture |

|
|||||
| Description | Import inner membrane translocase subunit TIM44, mitochondrial precursor. | |||||
|
IFIT2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.043778 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64112, Q62385 | Gene names | Ifit2, Garg39, Ifi54 | |||
|
Domain Architecture |
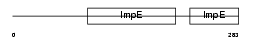
|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (Glucocorticoid- attenuated response gene 39 protein) (GARG-39). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.048868 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.030718 (rank : 170) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
NPHP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.025141 (rank : 174) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QY53, Q9D7G8, Q9QZP0, Q9WUZ2 | Gene names | Nphp1, Nph1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1. | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.038752 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
OLFL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.013947 (rank : 186) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BK62, Q3V2F2, Q8C1T0, Q99K37 | Gene names | Olfml3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactomedin-like protein 3 precursor. | |||||
|
PAX7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.005022 (rank : 191) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23759 | Gene names | PAX7, HUP1 | |||
|
Domain Architecture |
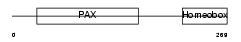
|
|||||
| Description | Paired box protein Pax-7 (HUP1). | |||||
|
PAX7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.005212 (rank : 190) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47239, Q9ES16 | Gene names | Pax7, Pax-7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-7. | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | 0.207464 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
APC7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.129046 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
SPAG5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.036353 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.034945 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
APC7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.122671 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.049103 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
LIN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.026688 (rank : 172) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.036802 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.022680 (rank : 179) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.013397 (rank : 187) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
CDC27_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.243097 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
DLG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.005378 (rank : 189) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
DLG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.005492 (rank : 188) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.046344 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.038903 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.032941 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.058284 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
ZN291_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.031009 (rank : 168) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
APXL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.025364 (rank : 173) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
CAR11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.035583 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BXL7, Q548H3 | Gene names | CARD11, CARMA1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 11 (CARD-containing MAGUK protein 3) (Carma 1). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.063105 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CCD91_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.036628 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.034658 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.015008 (rank : 183) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.014789 (rank : 185) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.038262 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.037327 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TPM3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.036005 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.030742 (rank : 169) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.048442 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.014980 (rank : 184) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.016132 (rank : 182) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
LIPB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.028867 (rank : 171) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.033503 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
MOES_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.023372 (rank : 177) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.031123 (rank : 167) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
NPHP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.053024 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.041328 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.039570 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
BBS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.241283 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |
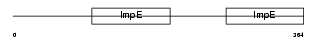
|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.237930 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |
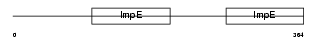
|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
CDC23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.064016 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CDC23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.058812 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.105921 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.106378 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
DNJB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050218 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |
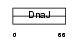
|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050659 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |
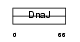
|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJC9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.053273 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
FKB11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.058424 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
Domain Architecture |
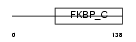
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.059429 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.093106 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.093473 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.087795 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
FKB1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.087854 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
FKBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.075607 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.076038 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.069105 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.065839 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.142269 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.140344 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |
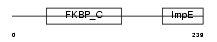
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.054484 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |
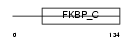
|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
IFIT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.050283 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09914, Q96QM5 | Gene names | IFIT1, G10P1, IFI56 | |||
|
Domain Architecture |
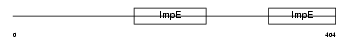
|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K). | |||||
|
IFIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.062025 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64282 | Gene names | Ifit1, Garg16, Ifi56 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K) (Glucocorticoid- attenuated response gene 16 protein) (GARG-16). | |||||
|
IFIT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.072011 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14879, Q99634, Q9BSK7 | Gene names | IFIT3, IFI60, IFIT4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (IFIT-4) (Interferon-induced 60 kDa protein) (IFI-60K) (ISG-60) (CIG49) (Retinoic acid-induced gene G protein) (RIG-G). | |||||
|
K0103_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.050320 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15006, Q8WUE1 | Gene names | KIAA0103 | |||
|
Domain Architecture |
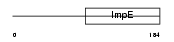
|
|||||
| Description | Tetratricopeptide repeat protein KIAA0103. | |||||
|
K0103_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.050045 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CRD2, Q925K1 | Gene names | Kiaa0103 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein KIAA0103. | |||||
|
NPHP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.053182 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
PEX5R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.084043 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
PEX5R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.084501 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
PEX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.099165 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
PEX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.104914 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
PPE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.050618 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14829, O15253, Q9NU21, Q9UJH0 | Gene names | PPEF1, PPEF, PPP7C | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 1 (EC 3.1.3.16) (PPEF-1) (Protein phosphatase with EF calcium-binding domain) (PPEF) (Serine/threonine-protein phosphatase 7) (PP7). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.081149 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.055778 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SUGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.290164 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
TTC15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.051222 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
TTC16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.251985 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.069696 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.070691 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.059160 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N0Z6 | Gene names | TTC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
TTC7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.054990 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BGB2, Q80XT5 | Gene names | Ttc7a, Ttc7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
ZCSL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.062588 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
ZCSL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.052063 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.055903 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.055603 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.98038e-156 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.912747 (rank : 2) | θ value | 5.99046e-137 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
OM34_MOUSE
|
||||||
| NC score | 0.807309 (rank : 3) | θ value | 1.52774e-15 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
OM34_HUMAN
|
||||||
| NC score | 0.805891 (rank : 4) | θ value | 3.07829e-16 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.780367 (rank : 5) | θ value | 4.02038e-16 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.777418 (rank : 6) | θ value | 2.35696e-16 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
TOM70_MOUSE
|
||||||
| NC score | 0.776716 (rank : 7) | θ value | 3.07829e-16 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.772626 (rank : 8) | θ value | 3.07829e-16 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
SGTB_HUMAN
|
||||||
| NC score | 0.746564 (rank : 9) | θ value | 1.133e-10 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
SGTB_MOUSE
|
||||||
| NC score | 0.745549 (rank : 10) | θ value | 1.47974e-10 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.731183 (rank : 11) | θ value | 1.2105e-12 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.728905 (rank : 12) | θ value | 2.69671e-12 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
SGTA_MOUSE
|
||||||
| NC score | 0.714025 (rank : 13) | θ value | 1.9326e-10 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
SGTA_HUMAN
|
||||||
| NC score | 0.711904 (rank : 14) | θ value | 1.9326e-10 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.711221 (rank : 15) | θ value | 1.25267e-09 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC12_MOUSE
|
||||||
| NC score | 0.702216 (rank : 16) | θ value | 6.21693e-09 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.698082 (rank : 17) | θ value | 6.43352e-06 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.692967 (rank : 18) | θ value | 1.29331e-14 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.688789 (rank : 19) | θ value | 1.09739e-05 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.671248 (rank : 20) | θ value | 9.90251e-15 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
UN45B_MOUSE
|
||||||
| NC score | 0.670852 (rank : 21) | θ value | 1.99529e-15 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
UN45B_HUMAN
|
||||||
| NC score | 0.668622 (rank : 22) | θ value | 5.8054e-15 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
TTC4_HUMAN
|
||||||
| NC score | 0.665979 (rank : 23) | θ value | 2.79066e-09 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
TTC4_MOUSE
|
||||||
| NC score | 0.647373 (rank : 24) | θ value | 2.79066e-09 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
DYXC1_MOUSE
|
||||||
| NC score | 0.628479 (rank : 25) | θ value | 8.40245e-06 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.611350 (rank : 26) | θ value | 2.21117e-06 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.585320 (rank : 27) | θ value | 1.25267e-09 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.582866 (rank : 28) | θ value | 4.44505e-15 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.580753 (rank : 29) | θ value | 4.44505e-15 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
F10A1_MOUSE
|
||||||
| NC score | 0.579938 (rank : 30) | θ value | 2.21117e-06 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.552974 (rank : 31) | θ value | 3.64472e-09 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.552153 (rank : 32) | θ value | 4.0297e-08 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
FKBP8_MOUSE
|
||||||
| NC score | 0.536280 (rank : 33) | θ value | 3.77169e-06 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.520028 (rank : 34) | θ value | 6.43352e-06 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
FKBP5_HUMAN
|
||||||
| NC score | 0.517356 (rank : 35) | θ value | 2.36244e-08 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP4_HUMAN
|
||||||
| NC score | 0.500812 (rank : 36) | θ value | 3.77169e-06 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_MOUSE
|
||||||
| NC score | 0.496683 (rank : 37) | θ value | 1.29631e-06 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
Z3H7B_HUMAN
|
||||||
| NC score | 0.494460 (rank : 38) | θ value | 0.00134147 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
FKBP4_MOUSE
|
||||||
| NC score | 0.492377 (rank : 39) | θ value | 4.92598e-06 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.451145 (rank : 40) | θ value | 0.0961366 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
PPP5_HUMAN
|
||||||
| NC score | 0.445137 (rank : 41) | θ value | 3.29651e-10 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
PPP5_MOUSE
|
||||||
| NC score | 0.444039 (rank : 42) | θ value | 1.9326e-10 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.430221 (rank : 43) | θ value | 1.80886e-08 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.422422 (rank : 44) | θ value | 6.21693e-09 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.417136 (rank : 45) | θ value | 0.00020696 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_MOUSE
|
||||||
| NC score | 0.403813 (rank : 46) | θ value | 0.000270298 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
Z3H7A_HUMAN
|
||||||
| NC score | 0.392369 (rank : 47) | θ value | 0.365318 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
AIP_HUMAN
|
||||||
| NC score | 0.386171 (rank : 48) | θ value | 0.00134147 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
WDTC1_MOUSE
|
||||||
| NC score | 0.384985 (rank : 49) | θ value | 1.99992e-07 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_HUMAN
|
||||||
| NC score | 0.383112 (rank : 50) | θ value | 5.81887e-07 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
SUGT1_MOUSE
|
||||||
| NC score | 0.376183 (rank : 51) | θ value | 0.47712 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
AIP_MOUSE
|
||||||
| NC score | 0.372597 (rank : 52) | θ value | 0.00509761 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.372558 (rank : 53) | θ value | 0.00020696 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
TTC13_HUMAN
|
||||||
| NC score | 0.365463 (rank : 54) | θ value | 0.0736092 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.349490 (rank : 55) | θ value | 0.000270298 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.337779 (rank : 56) | θ value | 3.19293e-05 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.325613 (rank : 57) | θ value | 0.00509761 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.321874 (rank : 58) | θ value | 0.0252991 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
SUGT1_HUMAN
|
||||||
| NC score | 0.290164 (rank : 59) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
K0372_HUMAN
|
||||||
| NC score | 0.269484 (rank : 60) | θ value | 0.279714 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.253605 (rank : 61) | θ value | 0.0113563 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
TTC16_MOUSE
|
||||||
| NC score | 0.251985 (rank : 62) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.246331 (rank : 63) | θ value | 0.0113563 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.243097 (rank : 64) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
BBS4_HUMAN
|
||||||
| NC score | 0.241283 (rank : 65) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| NC score | 0.237930 (rank : 66) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.237481 (rank : 67) | θ value | 0.00390308 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
IFT88_HUMAN
|
||||||
| NC score | 0.209167 (rank : 68) | θ value | 1.06291 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.207464 (rank : 69) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.202195 (rank : 70) | θ value | 0.21417 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
FKBP6_HUMAN
|
||||||
| NC score | 0.142269 (rank : 71) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_MOUSE
|
||||||
| NC score | 0.140344 (rank : 72) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
APC7_HUMAN
|
||||||
| NC score | 0.129046 (rank : 73) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
TTC8_HUMAN
|
||||||
| NC score | 0.128100 (rank : 74) | θ value | 0.0252991 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
APC7_MOUSE
|
||||||
| NC score | 0.122671 (rank : 75) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
TTC8_MOUSE
|
||||||
| NC score | 0.114220 (rank : 76) | θ value | 0.163984 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.106378 (rank : 77) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.105921 (rank : 78) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
PEX5_MOUSE
|
||||||
| NC score | 0.104914 (rank : 79) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
PEX5_HUMAN
|
||||||
| NC score | 0.099165 (rank : 80) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
BIN3_MOUSE
|
||||||
| NC score | 0.096714 (rank : 81) | θ value | 0.0252991 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JI08 | Gene names | Bin3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bridging integrator 3. | |||||
|
BIN3_HUMAN
|
||||||
| NC score | 0.094305 (rank : 82) | θ value | 0.21417 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQY0, Q9BVG2, Q9NVY9 | Gene names | BIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Bridging integrator 3. | |||||
|
FKB1A_MOUSE
|
||||||
| NC score | 0.093473 (rank : 83) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1A_HUMAN
|
||||||
| NC score | 0.093106 (rank : 84) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1B_MOUSE
|
||||||
| NC score | 0.087854 (rank : 85) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
FKB1B_HUMAN
|
||||||
| NC score | 0.087795 (rank : 86) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
PEX5R_MOUSE
|
||||||
| NC score | 0.084501 (rank : 87) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
PEX5R_HUMAN
|
||||||
| NC score | 0.084043 (rank : 88) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.081149 (rank : 89) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
FKBP2_MOUSE
|
||||||
| NC score | 0.076038 (rank : 90) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_HUMAN
|
||||||
| NC score | 0.075607 (rank : 91) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
IFIT3_HUMAN
|
||||||
| NC score | 0.072011 (rank : 92) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14879, Q99634, Q9BSK7 | Gene names | IFIT3, IFI60, IFIT4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (IFIT-4) (Interferon-induced 60 kDa protein) (IFI-60K) (ISG-60) (CIG49) (Retinoic acid-induced gene G protein) (RIG-G). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.070691 (rank : 93) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.069696 (rank : 94) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
FKBP3_HUMAN
|
||||||
| NC score | 0.069105 (rank : 95) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.068589 (rank : 96) | θ value | 0.0431538 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.067276 (rank : 97) | θ value | 0.0431538 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
FKBP3_MOUSE
|
||||||
| NC score | 0.065839 (rank : 98) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
CDC23_HUMAN
|
||||||
| NC score | 0.064016 (rank : 99) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.063552 (rank : 100) | θ value | 0.0330416 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.063105 (rank : 101) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
ZCSL3_HUMAN
|
||||||
| NC score | 0.062588 (rank : 102) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
IFIT1_MOUSE
|
||||||
| NC score | 0.062025 (rank : 103) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64282 | Gene names | Ifit1, Garg16, Ifi56 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K) (Glucocorticoid- attenuated response gene 16 protein) (GARG-16). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.061694 (rank : 104) | θ value | 0.365318 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.061019 (rank : 105) | θ value | 0.813845 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.059502 (rank : 106) | θ value | 0.21417 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
FKB11_MOUSE
|
||||||
| NC score | 0.059429 (rank : 107) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
TTC5_HUMAN
|
||||||
| NC score | 0.059160 (rank : 108) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N0Z6 | Gene names | TTC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.059052 (rank : 109) | θ value | 1.06291 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
CDC23_MOUSE
|
||||||
| NC score | 0.058812 (rank : 110) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
FKB11_HUMAN
|
||||||
| NC score | 0.058424 (rank : 111) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.058284 (rank : 112) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.055972 (rank : 113) | θ value | 0.00665767 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.055963 (rank : 114) | θ value | 0.0736092 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.055903 (rank : 115) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.055778 (rank : 116) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.055603 (rank : 117) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
TTC7A_MOUSE
|
||||||
| NC score | 0.054990 (rank : 118) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BGB2, Q80XT5 | Gene names | Ttc7a, Ttc7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
FKBP7_MOUSE
|
||||||
| NC score | 0.054484 (rank : 119) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.054240 (rank : 120) | θ value | 0.813845 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.054044 (rank : 121) | θ value | 0.813845 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.053670 (rank : 122) | θ value | 0.62314 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
DNJC9_MOUSE
|
||||||
| NC score | 0.053273 (rank : 123) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
NPHP3_HUMAN
|
||||||
| NC score | 0.053182 (rank : 124) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
NPHP3_MOUSE
|
||||||
| NC score | 0.053024 (rank : 125) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
ZCSL3_MOUSE
|
||||||
| NC score | 0.052063 (rank : 126) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
TTC15_MOUSE
|
||||||
| NC score | 0.051222 (rank : 127) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
DNJB8_MOUSE
|
||||||
| NC score | 0.050659 (rank : 128) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
PPE1_HUMAN
|
||||||
| NC score | 0.050618 (rank : 129) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14829, O15253, Q9NU21, Q9UJH0 | Gene names | PPEF1, PPEF, PPP7C | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 1 (EC 3.1.3.16) (PPEF-1) (Protein phosphatase with EF calcium-binding domain) (PPEF) (Serine/threonine-protein phosphatase 7) (PP7). | |||||
|
K0103_HUMAN
|
||||||
| NC score | 0.050320 (rank : 130) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15006, Q8WUE1 | Gene names | KIAA0103 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein KIAA0103. | |||||
|
IFIT1_HUMAN
|
||||||
| NC score | 0.050283 (rank : 131) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09914, Q96QM5 | Gene names | IFIT1, G10P1, IFI56 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K). | |||||
|
DNJB8_HUMAN
|
||||||
| NC score | 0.050218 (rank : 132) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
K0103_MOUSE
|
||||||
| NC score | 0.050045 (rank : 133) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CRD2, Q925K1 | Gene names | Kiaa0103 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein KIAA0103. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.049103 (rank : 134) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.048868 (rank : 135) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.048442 (rank : 136) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
NUF2_MOUSE
|
||||||
| NC score | 0.047837 (rank : 137) | θ value | 0.365318 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.047504 (rank : 138) | θ value | 0.21417 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.046344 (rank : 139) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
UBE4B_MOUSE
|
||||||
| NC score | 0.045506 (rank : 140) | θ value | 1.06291 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.044665 (rank : 141) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
FA50A_MOUSE
|
||||||
| NC score | 0.044430 (rank : 142) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WV03, Q9D866 | Gene names | Fam50a, D0HXS9928E, Xap5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM50A (XAP-5 protein). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.044425 (rank : 143) | θ value | 1.06291 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
IFIT2_MOUSE
|
||||||
| NC score | 0.043778 (rank : 144) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64112, Q62385 | Gene names | Ifit2, Garg39, Ifi54 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (Glucocorticoid- attenuated response gene 39 protein) (GARG-39). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.043010 (rank : 145) | θ value | 0.62314 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
KIF3A_HUMAN
|
||||||
| NC score | 0.042026 (rank : 146) | θ value | 0.279714 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.041566 (rank : 147) | θ value | 0.125558 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.041328 (rank : 148) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.039570 (rank : 149) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.038903 (rank : 150) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.038752 (rank : 151) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.038262 (rank : 152) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.037362 (rank : 153) | θ value | 0.813845 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.037327 (rank : 154) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.036802 (rank : 155) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CCD91_MOUSE
|
||||||
| NC score | 0.036628 (rank : 156) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
SPAG5_HUMAN
|
||||||
| NC score | 0.036353 (rank : 157) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
TPM3_HUMAN
|
||||||
| NC score | 0.036005 (rank : 158) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
CAR11_HUMAN
|
||||||
| NC score | 0.035583 (rank : 159) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BXL7, Q548H3 | Gene names | CARD11, CARMA1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 11 (CARD-containing MAGUK protein 3) (Carma 1). | |||||
|
STAP1_HUMAN
|
||||||
| NC score | 0.035549 (rank : 160) | θ value | 1.06291 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULZ2 | Gene names | STAP1, BRDG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1) (BCR downstream signaling protein 1) (Docking protein BRDG1). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.034945 (rank : 161) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.034658 (rank : 162) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
TIM44_MOUSE
|
||||||
| NC score | 0.034322 (rank : 163) | θ value | 1.38821 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35857 | Gene names | Timm44, Mimt44, Tim44 | |||
|
Domain Architecture |

|
|||||
| Description | Import inner membrane translocase subunit TIM44, mitochondrial precursor. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.033867 (rank : 164) | θ value | 0.365318 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.033503 (rank : 165) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.032941 (rank : 166) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.031123 (rank : 167) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
ZN291_HUMAN
|
||||||
| NC score | 0.031009 (rank : 168) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.030742 (rank : 169) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.030718 (rank : 170) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
LIPB2_MOUSE
|
||||||
| NC score | 0.028867 (rank : 171) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
LIN1_HUMAN
|
||||||
| NC score | 0.026688 (rank : 172) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.025364 (rank : 173) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
NPHP1_MOUSE
|
||||||
| NC score | 0.025141 (rank : 174) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QY53, Q9D7G8, Q9QZP0, Q9WUZ2 | Gene names | Nphp1, Nph1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1. | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.024638 (rank : 175) | θ value | 1.06291 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.023894 (rank : 176) | θ value | 0.00228821 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.023372 (rank : 177) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.023171 (rank : 178) | θ value | 1.38821 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.022680 (rank : 179) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.022626 (rank : 180) | θ value | 0.365318 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.020355 (rank : 181) | θ value | 1.38821 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.016132 (rank : 182) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.015008 (rank : 183) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.014980 (rank : 184) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.014789 (rank : 185) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
OLFL3_MOUSE
|
||||||
| NC score | 0.013947 (rank : 186) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BK62, Q3V2F2, Q8C1T0, Q99K37 | Gene names | Olfml3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactomedin-like protein 3 precursor. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.013397 (rank : 187) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
DLG1_MOUSE
|
||||||
| NC score | 0.005492 (rank : 188) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
DLG1_HUMAN
|
||||||
| NC score | 0.005378 (rank : 189) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
PAX7_MOUSE
|
||||||
| NC score | 0.005212 (rank : 190) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47239, Q9ES16 | Gene names | Pax7, Pax-7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-7. | |||||
|
PAX7_HUMAN
|
||||||
| NC score | 0.005022 (rank : 191) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23759 | Gene names | PAX7, HUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-7 (HUP1). | |||||