Please be patient as the page loads
|
AIPL1_HUMAN
|
||||||
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AIPL1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_MOUSE
|
||||||
| θ value | 2.33393e-165 (rank : 2) | NC score | 0.981744 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIP_MOUSE
|
||||||
| θ value | 1.3501e-96 (rank : 3) | NC score | 0.953213 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
AIP_HUMAN
|
||||||
| θ value | 3.00772e-96 (rank : 4) | NC score | 0.952843 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
FKBP5_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 5) | NC score | 0.474615 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP4_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 6) | NC score | 0.484442 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |
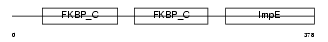
|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP4_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 7) | NC score | 0.477830 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 8) | NC score | 0.467933 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
OM34_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 9) | NC score | 0.491972 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 10) | NC score | 0.433223 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 11) | NC score | 0.366533 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 12) | NC score | 0.442191 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
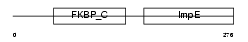
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 13) | NC score | 0.392525 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 14) | NC score | 0.362514 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
TTC12_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 15) | NC score | 0.414553 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |
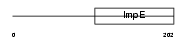
|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC12_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 16) | NC score | 0.413248 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
FKBP8_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 17) | NC score | 0.437146 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |
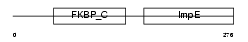
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
PPID_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 18) | NC score | 0.257575 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
TOM70_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 19) | NC score | 0.388004 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 20) | NC score | 0.375812 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TTC1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 21) | NC score | 0.417136 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
PPID_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 22) | NC score | 0.256138 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 23) | NC score | 0.407719 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 24) | NC score | 0.253871 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 25) | NC score | 0.308786 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.308310 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.249943 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.208650 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 29) | NC score | 0.209043 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
OM34_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 30) | NC score | 0.446455 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
UN45B_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 31) | NC score | 0.300637 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
STUB1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 32) | NC score | 0.317993 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 33) | NC score | 0.163741 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.032431 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.023519 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TTC16_MOUSE
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.183072 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
UN45B_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.295111 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
K0157_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.063328 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15018, Q96H11 | Gene names | KIAA0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.023055 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.021674 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
Z3H7B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.251151 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.047973 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
RUFY3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.035547 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
STUB1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.286339 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.017914 (rank : 134) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.018460 (rank : 133) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.028601 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.002658 (rank : 160) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PITM2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.024274 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
ZN202_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | -0.000857 (rank : 163) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95125, Q4JG21, Q9H1B9, Q9NSM4 | Gene names | ZNF202 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 202. | |||||
|
CD2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.035373 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.026816 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
K0157_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.056139 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TCJ1, Q3TB08, Q8K0R4 | Gene names | Kiaa0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.028745 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RUFY3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.032216 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.022303 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.010478 (rank : 150) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.021643 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.015162 (rank : 144) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.022606 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.017602 (rank : 138) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.022367 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
DHC24_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.054727 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15392, Q9HBA8 | Gene names | DHCR24, KIAA0018 | |||
|
Domain Architecture |
|
|||||
| Description | 24-dehydrocholesterol reductase precursor (EC 1.3.1.-) (3-beta- hydroxysterol delta-24-reductase) (Seladin-1) (Diminuto/dwarf1 homolog). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.034866 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.028774 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.017854 (rank : 136) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
INP5E_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.020116 (rank : 129) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
K0372_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.160507 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
MIS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.020634 (rank : 128) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03971, O75246 | Gene names | AMH, MIF | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.017586 (rank : 139) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PDLI7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.008177 (rank : 157) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.021261 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
SH22A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.020654 (rank : 127) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.019574 (rank : 131) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CCD16_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.023350 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
SGTA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.210668 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
SRTD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.025184 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.016567 (rank : 140) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.015665 (rank : 142) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.015436 (rank : 143) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.010918 (rank : 148) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.008327 (rank : 156) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.027789 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.017806 (rank : 137) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MK04_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | -0.000801 (rank : 161) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.024175 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.030158 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.018562 (rank : 132) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RPKL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.002936 (rank : 159) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R2S1, Q8R1D4 | Gene names | Rps6kl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase-like 1 (EC 2.7.11.1). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.009210 (rank : 151) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
UB7I1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.016485 (rank : 141) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.041736 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.031821 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.027107 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.008532 (rank : 154) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.019658 (rank : 130) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.011843 (rank : 146) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.017889 (rank : 135) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SGTA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.208064 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.008450 (rank : 155) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.022884 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | -0.000845 (rank : 162) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.008092 (rank : 158) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
GAB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.011695 (rank : 147) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.027117 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.027020 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.026496 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.027274 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.009041 (rank : 152) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.025458 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
XKR7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.008848 (rank : 153) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH72, Q9NUG5 | Gene names | XKR7, C20orf159, XRG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.014355 (rank : 145) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
TF2AY_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.010634 (rank : 149) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
BBS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.099870 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.097492 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
CDC27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.079036 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.210230 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
DYXC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.216738 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
F10A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.133918 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
F10A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.148552 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
F10A4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.150016 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
F10A5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.130701 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
FKB10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.068010 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.068929 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61576, Q8VHI1 | Gene names | Fkbp10, Fkbp-rs, Fkbp1-rs, Fkbp6, Fkbprp | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.080373 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
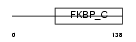
Domain Architecture |
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.082511 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
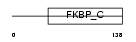
Domain Architecture |
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.063423 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NWM8 | Gene names | FKBP14, FKBP22 | |||
|
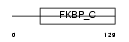
Domain Architecture |
|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (22 kDa FK506-binding protein) (FKBP-22). | |||||
|
FKB14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.069374 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59024 | Gene names | Fkbp14 | |||
|
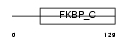
Domain Architecture |
|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKB1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.149273 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.149458 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |
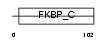
|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.120802 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
FKB1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.120578 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |
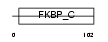
|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
FKBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.104966 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |
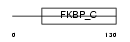
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.105563 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |
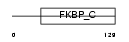
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.096782 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |
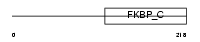
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.094775 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |
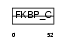
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.197637 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |
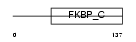
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.155224 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |
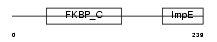
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.054998 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y680, Q86U65, Q96DA4, Q9Y6B0 | Gene names | FKBP7, FKBP23 | |||
|
Domain Architecture |
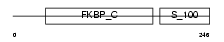
|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKBP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.076320 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |
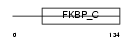
|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKBP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.067742 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95302, Q3MIR7, Q6IN76, Q6P2N1, Q96EX5, Q96IJ9 | Gene names | FKBP9, FKBP60, FKBP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKBP9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.069045 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z247, Q80ZZ6, Q8R386, Q9CVM0, Q9JHX5 | Gene names | Fkbp9, Fkbp60, Fkbp63 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP65RS). | |||||
|
IFT88_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.070813 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
IFT88_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.058648 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.100264 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
PPP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.140035 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
PPP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.140186 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
SGTB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.238911 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |
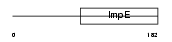
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
SGTB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.247398 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.063772 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.056325 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SUGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.094185 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |
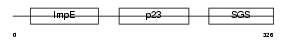
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
SUGT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.137664 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.056480 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
TTC13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.151071 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.110828 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
TTC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.088347 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
TTC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.207074 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
TTC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.201656 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
TTC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.159476 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
WDTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.086841 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.085523 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
Z3H7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.197752 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_MOUSE
|
||||||
| NC score | 0.981744 (rank : 2) | θ value | 2.33393e-165 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIP_MOUSE
|
||||||
| NC score | 0.953213 (rank : 3) | θ value | 1.3501e-96 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
AIP_HUMAN
|
||||||
| NC score | 0.952843 (rank : 4) | θ value | 3.00772e-96 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
OM34_HUMAN
|
||||||
| NC score | 0.491972 (rank : 5) | θ value | 5.81887e-07 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
FKBP4_HUMAN
|
||||||
| NC score | 0.484442 (rank : 6) | θ value | 4.30538e-10 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP4_MOUSE
|
||||||
| NC score | 0.477830 (rank : 7) | θ value | 7.34386e-10 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_HUMAN
|
||||||
| NC score | 0.474615 (rank : 8) | θ value | 8.67504e-11 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP5_MOUSE
|
||||||
| NC score | 0.467933 (rank : 9) | θ value | 6.21693e-09 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
OM34_MOUSE
|
||||||
| NC score | 0.446455 (rank : 10) | θ value | 0.0148317 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.442191 (rank : 11) | θ value | 2.44474e-05 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
FKBP8_MOUSE
|
||||||
| NC score | 0.437146 (rank : 12) | θ value | 9.29e-05 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.433223 (rank : 13) | θ value | 8.40245e-06 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.417136 (rank : 14) | θ value | 0.00020696 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC12_MOUSE
|
||||||
| NC score | 0.414553 (rank : 15) | θ value | 4.1701e-05 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.413248 (rank : 16) | θ value | 5.44631e-05 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.407719 (rank : 17) | θ value | 0.000461057 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.392525 (rank : 18) | θ value | 2.44474e-05 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.388004 (rank : 19) | θ value | 0.000121331 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_MOUSE
|
||||||
| NC score | 0.375812 (rank : 20) | θ value | 0.000121331 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.366533 (rank : 21) | θ value | 8.40245e-06 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.362514 (rank : 22) | θ value | 2.44474e-05 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.317993 (rank : 23) | θ value | 0.0563607 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.308786 (rank : 24) | θ value | 0.00869519 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.308310 (rank : 25) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
UN45B_HUMAN
|
||||||
| NC score | 0.300637 (rank : 26) | θ value | 0.0431538 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
UN45B_MOUSE
|
||||||
| NC score | 0.295111 (rank : 27) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.286339 (rank : 28) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.257575 (rank : 29) | θ value | 0.000121331 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.256138 (rank : 30) | θ value | 0.000461057 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.253871 (rank : 31) | θ value | 0.00869519 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
Z3H7B_HUMAN
|
||||||
| NC score | 0.251151 (rank : 32) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.249943 (rank : 33) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
SGTB_MOUSE
|
||||||
| NC score | 0.247398 (rank : 34) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
SGTB_HUMAN
|
||||||
| NC score | 0.238911 (rank : 35) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
DYXC1_MOUSE
|
||||||
| NC score | 0.216738 (rank : 36) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
SGTA_MOUSE
|
||||||
| NC score | 0.210668 (rank : 37) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.210230 (rank : 38) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.209043 (rank : 39) | θ value | 0.0148317 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.208650 (rank : 40) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
SGTA_HUMAN
|
||||||
| NC score | 0.208064 (rank : 41) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
TTC4_HUMAN
|
||||||
| NC score | 0.207074 (rank : 42) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
TTC4_MOUSE
|
||||||
| NC score | 0.201656 (rank : 43) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
Z3H7A_HUMAN
|
||||||
| NC score | 0.197752 (rank : 44) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
FKBP6_HUMAN
|
||||||
| NC score | 0.197637 (rank : 45) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
TTC16_MOUSE
|
||||||
| NC score | 0.183072 (rank : 46) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.163741 (rank : 47) | θ value | 0.0563607 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
K0372_HUMAN
|
||||||
| NC score | 0.160507 (rank : 48) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.159476 (rank : 49) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
FKBP6_MOUSE
|
||||||
| NC score | 0.155224 (rank : 50) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
TTC13_HUMAN
|
||||||
| NC score | 0.151071 (rank : 51) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.150016 (rank : 52) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
FKB1A_MOUSE
|
||||||
| NC score | 0.149458 (rank : 53) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1A_HUMAN
|
||||||
| NC score | 0.149273 (rank : 54) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
F10A1_MOUSE
|
||||||
| NC score | 0.148552 (rank : 55) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
PPP5_MOUSE
|
||||||
| NC score | 0.140186 (rank : 56) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
PPP5_HUMAN
|
||||||
| NC score | 0.140035 (rank : 57) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
SUGT1_MOUSE
|
||||||
| NC score | 0.137664 (rank : 58) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.133918 (rank : 59) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.130701 (rank : 60) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
FKB1B_HUMAN
|
||||||
| NC score | 0.120802 (rank : 61) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
FKB1B_MOUSE
|
||||||
| NC score | 0.120578 (rank : 62) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.110828 (rank : 63) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
FKBP2_MOUSE
|
||||||
| NC score | 0.105563 (rank : 64) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_HUMAN
|
||||||
| NC score | 0.104966 (rank : 65) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.100264 (rank : 66) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
BBS4_HUMAN
|
||||||
| NC score | 0.099870 (rank : 67) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| NC score | 0.097492 (rank : 68) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
FKBP3_HUMAN
|
||||||
| NC score | 0.096782 (rank : 69) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP3_MOUSE
|
||||||
| NC score | 0.094775 (rank : 70) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
SUGT1_HUMAN
|
||||||
| NC score | 0.094185 (rank : 71) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.088347 (rank : 72) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
WDTC1_HUMAN
|
||||||
| NC score | 0.086841 (rank : 73) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_MOUSE
|
||||||
| NC score | 0.085523 (rank : 74) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
FKB11_MOUSE
|
||||||
| NC score | 0.082511 (rank : 75) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB11_HUMAN
|
||||||
| NC score | 0.080373 (rank : 76) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.079036 (rank : 77) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
FKBP7_MOUSE
|
||||||
| NC score | 0.076320 (rank : 78) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
IFT88_HUMAN
|
||||||
| NC score | 0.070813 (rank : 79) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
FKB14_MOUSE
|
||||||
| NC score | 0.069374 (rank : 80) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59024 | Gene names | Fkbp14 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKBP9_MOUSE
|
||||||
| NC score | 0.069045 (rank : 81) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z247, Q80ZZ6, Q8R386, Q9CVM0, Q9JHX5 | Gene names | Fkbp9, Fkbp60, Fkbp63 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP65RS). | |||||
|
FKB10_MOUSE
|
||||||
| NC score | 0.068929 (rank : 82) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61576, Q8VHI1 | Gene names | Fkbp10, Fkbp-rs, Fkbp1-rs, Fkbp6, Fkbprp | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB10_HUMAN
|
||||||
| NC score | 0.068010 (rank : 83) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKBP9_HUMAN
|
||||||
| NC score | 0.067742 (rank : 84) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95302, Q3MIR7, Q6IN76, Q6P2N1, Q96EX5, Q96IJ9 | Gene names | FKBP9, FKBP60, FKBP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.063772 (rank : 85) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
FKB14_HUMAN
|
||||||
| NC score | 0.063423 (rank : 86) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NWM8 | Gene names | FKBP14, FKBP22 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (22 kDa FK506-binding protein) (FKBP-22). | |||||
|
K0157_HUMAN
|
||||||
| NC score | 0.063328 (rank : 87) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15018, Q96H11 | Gene names | KIAA0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.058648 (rank : 88) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.056480 (rank : 89) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.056325 (rank : 90) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
K0157_MOUSE
|
||||||
| NC score | 0.056139 (rank : 91) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TCJ1, Q3TB08, Q8K0R4 | Gene names | Kiaa0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
FKBP7_HUMAN
|
||||||
| NC score | 0.054998 (rank : 92) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y680, Q86U65, Q96DA4, Q9Y6B0 | Gene names | FKBP7, FKBP23 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
DHC24_HUMAN
|
||||||
| NC score | 0.054727 (rank : 93) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15392, Q9HBA8 | Gene names | DHCR24, KIAA0018 | |||
|
Domain Architecture |

|
|||||
| Description | 24-dehydrocholesterol reductase precursor (EC 1.3.1.-) (3-beta- hydroxysterol delta-24-reductase) (Seladin-1) (Diminuto/dwarf1 homolog). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.047973 (rank : 94) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.041736 (rank : 95) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
RUFY3_MOUSE
|
||||||
| NC score | 0.035547 (rank : 96) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.035373 (rank : 97) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.034866 (rank : 98) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.032431 (rank : 99) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RUFY3_HUMAN
|
||||||
| NC score | 0.032216 (rank : 100) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.031821 (rank : 101) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.030158 (rank : 102) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.028774 (rank : 103) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.028745 (rank : 104) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.028601 (rank : 105) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.027789 (rank : 106) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.027274 (rank : 107) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.027117 (rank : 108) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.027107 (rank : 109) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.027020 (rank : 110) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.026816 (rank : 111) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.026496 (rank : 112) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.025458 (rank : 113) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
SRTD2_HUMAN
|
||||||
| NC score | 0.025184 (rank : 114) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
PITM2_MOUSE
|
||||||
| NC score | 0.024274 (rank : 115) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.024175 (rank : 116) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.023519 (rank : 117) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CCD16_MOUSE
|
||||||
| NC score | 0.023350 (rank : 118) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.023055 (rank : 119) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.022884 (rank : 120) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.022606 (rank : 121) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.022367 (rank : 122) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.022303 (rank : 123) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.021674 (rank : 124) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.021643 (rank : 125) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.021261 (rank : 126) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
SH22A_MOUSE
|
||||||
| NC score | 0.020654 (rank : 127) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
MIS_HUMAN
|
||||||
| NC score | 0.020634 (rank : 128) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03971, O75246 | Gene names | AMH, MIF | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
INP5E_HUMAN
|
||||||
| NC score | 0.020116 (rank : 129) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.019658 (rank : 130) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.019574 (rank : 131) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.018562 (rank : 132) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.018460 (rank : 133) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.017914 (rank : 134) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.017889 (rank : 135) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.017854 (rank : 136) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.017806 (rank : 137) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.017602 (rank : 138) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.017586 (rank : 139) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.016567 (rank : 140) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
UB7I1_HUMAN
|
||||||
| NC score | 0.016485 (rank : 141) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.015665 (rank : 142) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.015436 (rank : 143) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.015162 (rank : 144) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.014355 (rank : 145) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.011843 (rank : 146) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.011695 (rank : 147) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.010918 (rank : 148) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
TF2AY_MOUSE
|
||||||
| NC score | 0.010634 (rank : 149) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.010478 (rank : 150) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.009210 (rank : 151) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.009041 (rank : 152) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
XKR7_HUMAN
|
||||||
| NC score | 0.008848 (rank : 153) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH72, Q9NUG5 | Gene names | XKR7, C20orf159, XRG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.008532 (rank : 154) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.008450 (rank : 155) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.008327 (rank : 156) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
PDLI7_MOUSE
|
||||||
| NC score | 0.008177 (rank : 157) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.008092 (rank : 158) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
RPKL1_MOUSE
|
||||||
| NC score | 0.002936 (rank : 159) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R2S1, Q8R1D4 | Gene names | Rps6kl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase-like 1 (EC 2.7.11.1). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.002658 (rank : 160) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
MK04_MOUSE
|
||||||
| NC score | -0.000801 (rank : 161) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | -0.000845 (rank : 162) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZN202_HUMAN
|
||||||
| NC score | -0.000857 (rank : 163) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95125, Q4JG21, Q9H1B9, Q9NSM4 | Gene names | ZNF202 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 202. | |||||