Please be patient as the page loads
|
K0157_HUMAN
|
||||||
| SwissProt Accessions | Q15018, Q96H11 | Gene names | KIAA0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K0157_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q15018, Q96H11 | Gene names | KIAA0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
K0157_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.971878 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q3TCJ1, Q3TB08, Q8K0R4 | Gene names | Kiaa0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
TRI45_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 3) | NC score | 0.135457 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PFY8, Q8BVT5 | Gene names | Trim45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45. | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 4) | NC score | 0.094905 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
TRI45_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 5) | NC score | 0.125326 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8W5, Q5T2K4, Q5T2K5, Q8IYV6 | Gene names | TRIM45, RNF99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45 (RING finger protein 99). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 6) | NC score | 0.063886 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.016131 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.057211 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
CING_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.062537 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.063328 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
DBNL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.062255 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
THAP7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.111223 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP7_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.111317 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.015782 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
SNIP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.090933 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
CING_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.059418 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
DAAM1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.068981 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.075921 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.053800 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
FA13A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.041138 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
C102B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.071261 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
DAAM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.063450 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
ABI1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.037410 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP0, O15147, O76049, O95060, Q5W070, Q5W072, Q8TB63, Q96S81, Q9NXZ9, Q9NYB8 | Gene names | ABI1, SSH3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Nap1-binding protein) (Nap1BP) (Abl-binding protein 4) (AblBP4). | |||||
|
ABI1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.038361 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CBW3, Q60747, Q91ZM5, Q923I9, Q99KH4 | Gene names | Abi1, Ssh3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Ablphilin-1). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.048162 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
DAAM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.057291 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.049813 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.013343 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.032661 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
TEX9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.073280 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
CA036_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.058613 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BRE0, Q9CRS3, Q9JMF2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf36 homolog. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.057794 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
GP156_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.056753 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
PDXL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.043659 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.056407 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.037762 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
CEP57_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.059724 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86XR8, Q14704, Q8IXP0, Q9BVF9 | Gene names | CEP57, KIAA0092, TSP57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin) (FGF2-interacting protein). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.045089 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.041086 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
BRCA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.041576 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.058897 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
EFS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.026352 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
FZD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.013611 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIP6, Q810M0, Q9JIP5, Q9WUJ2 | Gene names | Fzd2, Fzd10 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-2 precursor (Fz-2) (mFz2) (mFz10). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.017800 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
ZF106_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.020326 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.048086 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.021504 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.024964 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
POLS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.030944 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.042125 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
IL6RA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.020495 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08887, Q16202, Q53EQ7, Q5FWG2, Q5VZ23 | Gene names | IL6R | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor alpha chain precursor (IL-6R-alpha) (IL-6R 1) (Membrane glycoprotein 80) (gp80) (CD126 antigen). | |||||
|
KIF17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.028195 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2E2, O95077, Q53YS6, Q5VWA9, Q6GSA8, Q8N411 | Gene names | KIF17, KIAA1405, KIF3X | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF17 (KIF3-related motor protein). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.037463 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.041520 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.040201 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.024744 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TEX9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.058541 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.042398 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
CM007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.021711 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
DYR1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.005027 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y463, O75258, O75788, O75789 | Gene names | DYRK1B, MIRK | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1) (Mirk protein kinase) (Minibrain-related kinase). | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.031541 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
KIF3B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.029136 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
M3K12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.004662 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.041801 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.034577 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.020357 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.045635 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.046249 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.011397 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
KIF3B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.027839 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
KIF3C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.026971 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35066, O35229 | Gene names | Kif3c | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.028081 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.024427 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
SPAG5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.045999 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
SPN90_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.017559 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESJ4, Q68G72 | Gene names | Nckipsd, Spin90, Wasbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (N-WASP-interacting protein, 90kDa) (Wiskott-Aldrich syndrome protein-binding protein WISH) (N-WASP-binding protein). | |||||
|
SURF6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.046357 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70279, Q8BPI4, Q8BRL7, Q8R0Q7 | Gene names | Surf6, Surf-6 | |||
|
Domain Architecture |
|
|||||
| Description | Surfeit locus protein 6. | |||||
|
ZN263_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | -0.001532 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14978, O43387, Q96H95 | Gene names | ZNF263, FPM315 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 263 (Zinc finger protein FPM315). | |||||
|
CBLB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.010335 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.039930 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.036117 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.041645 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FZD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.010687 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14332 | Gene names | FZD2 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-2 precursor (Fz-2) (hFz2) (FzE2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.039100 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.029691 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KIF3C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.025270 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.026879 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
LAMC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.015501 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LIPB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.027526 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.028952 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.016661 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.008568 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
RABE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.043578 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |
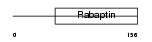
|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.033247 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.033251 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
TF3C2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.019031 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.008834 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.005311 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
HOOK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050383 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
K0157_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q15018, Q96H11 | Gene names | KIAA0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
K0157_MOUSE
|
||||||
| NC score | 0.971878 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q3TCJ1, Q3TB08, Q8K0R4 | Gene names | Kiaa0157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0157. | |||||
|
TRI45_MOUSE
|
||||||
| NC score | 0.135457 (rank : 3) | θ value | 0.00035302 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PFY8, Q8BVT5 | Gene names | Trim45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45. | |||||
|
TRI45_HUMAN
|
||||||
| NC score | 0.125326 (rank : 4) | θ value | 0.00298849 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8W5, Q5T2K4, Q5T2K5, Q8IYV6 | Gene names | TRIM45, RNF99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45 (RING finger protein 99). | |||||
|
THAP7_MOUSE
|
||||||
| NC score | 0.111317 (rank : 5) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP7_HUMAN
|
||||||
| NC score | 0.111223 (rank : 6) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.094905 (rank : 7) | θ value | 0.00134147 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
SNIP_HUMAN
|
||||||
| NC score | 0.090933 (rank : 8) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.075921 (rank : 9) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
TEX9_MOUSE
|
||||||
| NC score | 0.073280 (rank : 10) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
C102B_HUMAN
|
||||||
| NC score | 0.071261 (rank : 11) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
DAAM1_HUMAN
|
||||||
| NC score | 0.068981 (rank : 12) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.063886 (rank : 13) | θ value | 0.0148317 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DAAM1_MOUSE
|
||||||
| NC score | 0.063450 (rank : 14) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.063328 (rank : 15) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.062537 (rank : 16) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
DBNL_MOUSE
|
||||||
| NC score | 0.062255 (rank : 17) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
CEP57_HUMAN
|
||||||
| NC score | 0.059724 (rank : 18) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86XR8, Q14704, Q8IXP0, Q9BVF9 | Gene names | CEP57, KIAA0092, TSP57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin) (FGF2-interacting protein). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.059418 (rank : 19) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.058897 (rank : 20) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
CA036_MOUSE
|
||||||
| NC score | 0.058613 (rank : 21) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BRE0, Q9CRS3, Q9JMF2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf36 homolog. | |||||
|
TEX9_HUMAN
|
||||||
| NC score | 0.058541 (rank : 22) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.057794 (rank : 23) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
DAAM2_HUMAN
|
||||||
| NC score | 0.057291 (rank : 24) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.057211 (rank : 25) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
GP156_HUMAN
|
||||||
| NC score | 0.056753 (rank : 26) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.056407 (rank : 27) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.053800 (rank : 28) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
HOOK2_MOUSE
|
||||||
| NC score | 0.050383 (rank : 29) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.049813 (rank : 30) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.048162 (rank : 31) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.048086 (rank : 32) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
SURF6_MOUSE
|
||||||
| NC score | 0.046357 (rank : 33) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70279, Q8BPI4, Q8BRL7, Q8R0Q7 | Gene names | Surf6, Surf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.046249 (rank : 34) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
SPAG5_HUMAN
|
||||||
| NC score | 0.045999 (rank : 35) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.045635 (rank : 36) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.045089 (rank : 37) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
PDXL2_HUMAN
|
||||||
| NC score | 0.043659 (rank : 38) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
RABE2_HUMAN
|
||||||
| NC score | 0.043578 (rank : 39) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.042398 (rank : 40) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.042125 (rank : 41) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.041801 (rank : 42) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.041645 (rank : 43) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
BRCA2_MOUSE
|
||||||
| NC score | 0.041576 (rank : 44) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.041520 (rank : 45) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.041138 (rank : 46) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.041086 (rank : 47) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.040201 (rank : 48) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.039930 (rank : 49) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.039100 (rank : 50) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
ABI1_MOUSE
|
||||||
| NC score | 0.038361 (rank : 51) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CBW3, Q60747, Q91ZM5, Q923I9, Q99KH4 | Gene names | Abi1, Ssh3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Ablphilin-1). | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.037762 (rank : 52) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.037463 (rank : 53) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
ABI1_HUMAN
|
||||||
| NC score | 0.037410 (rank : 54) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP0, O15147, O76049, O95060, Q5W070, Q5W072, Q8TB63, Q96S81, Q9NXZ9, Q9NYB8 | Gene names | ABI1, SSH3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Nap1-binding protein) (Nap1BP) (Abl-binding protein 4) (AblBP4). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.036117 (rank : 55) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.034577 (rank : 56) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.033251 (rank : 57) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.033247 (rank : 58) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.032661 (rank : 59) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.031541 (rank : 60) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
POLS_MOUSE
|
||||||
| NC score | 0.030944 (rank : 61) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.029691 (rank : 62) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KIF3B_HUMAN
|
||||||
| NC score | 0.029136 (rank : 63) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.028952 (rank : 64) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
KIF17_HUMAN
|
||||||
| NC score | 0.028195 (rank : 65) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2E2, O95077, Q53YS6, Q5VWA9, Q6GSA8, Q8N411 | Gene names | KIF17, KIAA1405, KIF3X | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (KIF3-related motor protein). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.028081 (rank : 66) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
KIF3B_MOUSE
|
||||||
| NC score | 0.027839 (rank : 67) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
LIPB1_HUMAN
|
||||||
| NC score | 0.027526 (rank : 68) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
KIF3C_MOUSE
|
||||||
| NC score | 0.026971 (rank : 69) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35066, O35229 | Gene names | Kif3c | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.026879 (rank : 70) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.026352 (rank : 71) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
KIF3C_HUMAN
|
||||||
| NC score | 0.025270 (rank : 72) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.024964 (rank : 73) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.024744 (rank : 74) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.024427 (rank : 75) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.021711 (rank : 76) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.021504 (rank : 77) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
IL6RA_HUMAN
|
||||||
| NC score | 0.020495 (rank : 78) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08887, Q16202, Q53EQ7, Q5FWG2, Q5VZ23 | Gene names | IL6R | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor alpha chain precursor (IL-6R-alpha) (IL-6R 1) (Membrane glycoprotein 80) (gp80) (CD126 antigen). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.020357 (rank : 79) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
ZF106_HUMAN
|
||||||
| NC score | 0.020326 (rank : 80) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
TF3C2_HUMAN
|
||||||
| NC score | 0.019031 (rank : 81) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.017800 (rank : 82) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
SPN90_MOUSE
|
||||||
| NC score | 0.017559 (rank : 83) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESJ4, Q68G72 | Gene names | Nckipsd, Spin90, Wasbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (N-WASP-interacting protein, 90kDa) (Wiskott-Aldrich syndrome protein-binding protein WISH) (N-WASP-binding protein). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.016661 (rank : 84) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.016131 (rank : 85) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.015782 (rank : 86) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
LAMC1_MOUSE
|
||||||
| NC score | 0.015501 (rank : 87) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
FZD2_MOUSE
|
||||||
| NC score | 0.013611 (rank : 88) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIP6, Q810M0, Q9JIP5, Q9WUJ2 | Gene names | Fzd2, Fzd10 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-2 precursor (Fz-2) (mFz2) (mFz10). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.013343 (rank : 89) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.011397 (rank : 90) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
FZD2_HUMAN
|
||||||
| NC score | 0.010687 (rank : 91) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14332 | Gene names | FZD2 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-2 precursor (Fz-2) (hFz2) (FzE2). | |||||
|
CBLB_MOUSE
|
||||||
| NC score | 0.010335 (rank : 92) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.008834 (rank : 93) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.008568 (rank : 94) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
ULK1_HUMAN
|
||||||
| NC score | 0.005311 (rank : 95) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
DYR1B_HUMAN
|
||||||
| NC score | 0.005027 (rank : 96) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y463, O75258, O75788, O75789 | Gene names | DYRK1B, MIRK | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1) (Mirk protein kinase) (Minibrain-related kinase). | |||||
|
M3K12_MOUSE
|
||||||
| NC score | 0.004662 (rank : 97) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
ZN263_HUMAN
|
||||||
| NC score | -0.001532 (rank : 98) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14978, O43387, Q96H95 | Gene names | ZNF263, FPM315 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 263 (Zinc finger protein FPM315). | |||||