Please be patient as the page loads
|
TTC12_HUMAN
|
||||||
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TTC12_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC12_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.981012 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
SGTB_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 3) | NC score | 0.728901 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 4) | NC score | 0.695393 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 5) | NC score | 0.697450 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
SGTB_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 6) | NC score | 0.729201 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 7) | NC score | 0.665340 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
SGTA_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 8) | NC score | 0.698163 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
OM34_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 9) | NC score | 0.743992 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 10) | NC score | 0.702853 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
OM34_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 11) | NC score | 0.741788 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 12) | NC score | 0.695524 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
SGTA_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 13) | NC score | 0.690052 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |
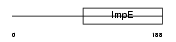
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
STUB1_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 14) | NC score | 0.706634 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
FKBP5_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 15) | NC score | 0.529040 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
STUB1_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 16) | NC score | 0.694198 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
PPP5_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 17) | NC score | 0.415796 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
FKBP4_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 18) | NC score | 0.509640 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |
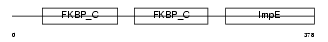
|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
PPP5_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 19) | NC score | 0.414426 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
FKBP4_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 20) | NC score | 0.502947 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 21) | NC score | 0.515011 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
TTC1_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 22) | NC score | 0.711221 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 23) | NC score | 0.499774 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 24) | NC score | 0.505925 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
TOM70_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 25) | NC score | 0.666460 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 26) | NC score | 0.670800 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 27) | NC score | 0.572886 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
TTC4_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 28) | NC score | 0.602303 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |
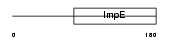
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
UN45B_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 29) | NC score | 0.539094 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
WDTC1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 30) | NC score | 0.375066 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
TTC4_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 31) | NC score | 0.615177 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |
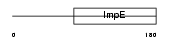
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
WDTC1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 32) | NC score | 0.371534 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
Z3H7B_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 33) | NC score | 0.501035 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 34) | NC score | 0.563407 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
UN45B_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 35) | NC score | 0.531852 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
F10A4_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 36) | NC score | 0.504773 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 37) | NC score | 0.542416 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
AIP_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 38) | NC score | 0.414462 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
DYXC1_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 39) | NC score | 0.582413 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
AIP_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 40) | NC score | 0.403265 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
F10A1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 41) | NC score | 0.467267 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 42) | NC score | 0.356358 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 43) | NC score | 0.413248 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
F10A5_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 44) | NC score | 0.468057 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
AIPL1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 45) | NC score | 0.403184 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
TTC3_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 46) | NC score | 0.315592 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
F10A1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 47) | NC score | 0.489100 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
PPID_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 48) | NC score | 0.317923 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPID_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 49) | NC score | 0.302442 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
TPM3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 50) | NC score | 0.065889 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
TPM3_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 51) | NC score | 0.066469 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
Z3H7A_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 52) | NC score | 0.389092 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
BBS4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 53) | NC score | 0.287030 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
FKBP6_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 54) | NC score | 0.200203 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 55) | NC score | 0.231513 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 56) | NC score | 0.224224 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 57) | NC score | 0.304853 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
BBS4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 58) | NC score | 0.282034 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
CTNB1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 59) | NC score | 0.057021 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.057037 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
K0372_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 61) | NC score | 0.273019 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 62) | NC score | 0.310644 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
FKBP6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 63) | NC score | 0.194755 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |
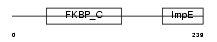
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 0.163984 (rank : 64) | NC score | 0.212378 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
IFT88_HUMAN
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.225553 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 66) | NC score | 0.208004 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.415546 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
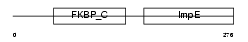
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
FKBP8_MOUSE
|
||||||
| θ value | 0.365318 (rank : 68) | NC score | 0.425244 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |
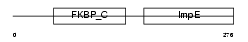
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 0.365318 (rank : 69) | NC score | 0.023476 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 70) | NC score | 0.026629 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
GPSM2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.100569 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.102259 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
IFT88_MOUSE
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.206776 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.47712 (rank : 74) | NC score | 0.026583 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.47712 (rank : 75) | NC score | 0.025992 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.026849 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.046278 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
CDC27_HUMAN
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.245875 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.022022 (rank : 142) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
TPM4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.054664 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P67936, P07226, Q15659, Q9BU85, Q9H8Q3 | Gene names | TPM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4) (TM30p1). | |||||
|
TTC13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.319198 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 0.813845 (rank : 82) | NC score | 0.130061 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.021621 (rank : 143) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.018188 (rank : 147) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.021291 (rank : 144) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
GPSM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.088751 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86YR5, Q86SR5, Q969T1, Q9UFS8 | Gene names | GPSM1, AGS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
GPSM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.098623 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.129152 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.391441 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
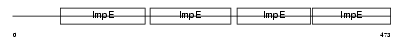
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
ZMY12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.036945 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.036587 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
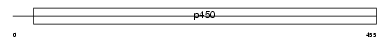
CP39A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.009570 (rank : 159) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.020137 (rank : 145) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TPM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.048086 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.048409 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.011870 (rank : 156) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CDC23_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.087802 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CDC23_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.085398 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.026732 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CING_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.024436 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
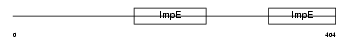
IFIT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.052731 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09914, Q96QM5 | Gene names | IFIT1, G10P1, IFI56 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K). | |||||
|
INSC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.022801 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q1MX18, Q1MX19, Q3C1V6, Q4AC95, Q4AC96, Q4AC97, Q4AC98 | Gene names | INSC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inscuteable homolog. | |||||
|
SIL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.029138 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EPK6, Q91V34 | Gene names | Sil1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor. | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.013309 (rank : 155) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.011131 (rank : 158) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.016865 (rank : 151) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.039305 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
RB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.011691 (rank : 157) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06400, P78499, Q5VW46, Q8IZL4 | Gene names | RB1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-associated protein (PP110) (P105-RB) (RB). | |||||
|
RPTOR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.023659 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N122, Q8N4V9, Q8TB32, Q9P2P3 | Gene names | RAPTOR, KIAA1303 | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
SIL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.028643 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H173, Q8N2L3 | Gene names | SIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor (BiP-associated protein) (BAP). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.019088 (rank : 146) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TPM4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.043885 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
TSH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.017300 (rank : 149) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
TSH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.017708 (rank : 148) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.023159 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
HS70L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.003373 (rank : 161) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.015993 (rank : 153) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.008002 (rank : 160) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
RBP22_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.038442 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RGPD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.037728 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.025709 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
RPTOR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.022761 (rank : 141) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K4Q0, Q8C9W9, Q8CBY4, Q8CDY8, Q9D4H3 | Gene names | Raptor | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.017028 (rank : 150) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.014934 (rank : 154) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
TSH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.016174 (rank : 152) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
APC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.084625 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
APC7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.082999 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.092407 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.093273 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
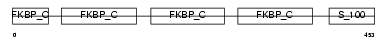
FKB10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.061882 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.061345 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61576, Q8VHI1 | Gene names | Fkbp10, Fkbp-rs, Fkbp1-rs, Fkbp6, Fkbprp | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
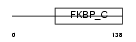
FKB11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.069454 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.071805 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.052148 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NWM8 | Gene names | FKBP14, FKBP22 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (22 kDa FK506-binding protein) (FKBP-22). | |||||
|
FKB14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.055416 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59024 | Gene names | Fkbp14 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKB1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.110332 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.109491 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.105328 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
FKB1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.104857 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
FKBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.091520 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.092038 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.088272 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.087327 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |
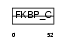
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.066914 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |
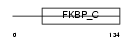
|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKBP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.059983 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95302, Q3MIR7, Q6IN76, Q6P2N1, Q96EX5, Q96IJ9 | Gene names | FKBP9, FKBP60, FKBP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKBP9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.060823 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z247, Q80ZZ6, Q8R386, Q9CVM0, Q9JHX5 | Gene names | Fkbp9, Fkbp60, Fkbp63 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP65RS). | |||||
|
IFIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.050006 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64282 | Gene names | Ifit1, Garg16, Ifi56 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K) (Glucocorticoid- attenuated response gene 16 protein) (GARG-16). | |||||
|
IFIT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.064392 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14879, Q99634, Q9BSK7 | Gene names | IFIT3, IFI60, IFIT4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (IFIT-4) (Interferon-induced 60 kDa protein) (IFI-60K) (ISG-60) (CIG49) (Retinoic acid-induced gene G protein) (RIG-G). | |||||
|
PEX5R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.078631 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
PEX5R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.079913 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
PEX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.101828 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
PEX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.105088 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
RAPSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.062201 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13702, Q9BTD9 | Gene names | RAPSN | |||
|
Domain Architecture |
|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
RAPSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.056905 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |
|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
SUGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.269651 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
SUGT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.332534 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
TTC14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.224600 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
TTC16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.263329 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TTC7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.056123 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGB2, Q80XT5 | Gene names | Ttc7a, Ttc7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
TTC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.065455 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
TTC8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.060950 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC12_MOUSE
|
||||||
| NC score | 0.981012 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
OM34_HUMAN
|
||||||
| NC score | 0.743992 (rank : 3) | θ value | 8.38298e-14 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
OM34_MOUSE
|
||||||
| NC score | 0.741788 (rank : 4) | θ value | 2.43908e-13 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
SGTB_HUMAN
|
||||||
| NC score | 0.729201 (rank : 5) | θ value | 2.20605e-14 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
SGTB_MOUSE
|
||||||
| NC score | 0.728901 (rank : 6) | θ value | 5.8054e-15 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.711221 (rank : 7) | θ value | 1.25267e-09 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.706634 (rank : 8) | θ value | 3.89403e-11 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.702853 (rank : 9) | θ value | 8.38298e-14 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SGTA_MOUSE
|
||||||
| NC score | 0.698163 (rank : 10) | θ value | 6.41864e-14 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.697450 (rank : 11) | θ value | 1.68911e-14 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.695524 (rank : 12) | θ value | 9.26847e-13 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.695393 (rank : 13) | θ value | 1.29331e-14 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.694198 (rank : 14) | θ value | 1.9326e-10 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
SGTA_HUMAN
|
||||||
| NC score | 0.690052 (rank : 15) | θ value | 4.59992e-12 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
TOM70_MOUSE
|
||||||
| NC score | 0.670800 (rank : 16) | θ value | 2.13673e-09 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.666460 (rank : 17) | θ value | 2.13673e-09 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.665340 (rank : 18) | θ value | 2.88119e-14 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC4_HUMAN
|
||||||
| NC score | 0.615177 (rank : 19) | θ value | 4.45536e-07 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
TTC4_MOUSE
|
||||||
| NC score | 0.602303 (rank : 20) | θ value | 8.97725e-08 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
DYXC1_MOUSE
|
||||||
| NC score | 0.582413 (rank : 21) | θ value | 4.92598e-06 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.572886 (rank : 22) | θ value | 4.0297e-08 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.563407 (rank : 23) | θ value | 1.29631e-06 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.542416 (rank : 24) | θ value | 2.21117e-06 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
UN45B_MOUSE
|
||||||
| NC score | 0.539094 (rank : 25) | θ value | 2.61198e-07 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
UN45B_HUMAN
|
||||||
| NC score | 0.531852 (rank : 26) | θ value | 1.29631e-06 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
FKBP5_HUMAN
|
||||||
| NC score | 0.529040 (rank : 27) | θ value | 8.67504e-11 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP5_MOUSE
|
||||||
| NC score | 0.515011 (rank : 28) | θ value | 5.62301e-10 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
FKBP4_HUMAN
|
||||||
| NC score | 0.509640 (rank : 29) | θ value | 4.30538e-10 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.505925 (rank : 30) | θ value | 2.13673e-09 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.504773 (rank : 31) | θ value | 1.69304e-06 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
FKBP4_MOUSE
|
||||||
| NC score | 0.502947 (rank : 32) | θ value | 5.62301e-10 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
Z3H7B_HUMAN
|
||||||
| NC score | 0.501035 (rank : 33) | θ value | 9.92553e-07 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.499774 (rank : 34) | θ value | 2.13673e-09 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
F10A1_MOUSE
|
||||||
| NC score | 0.489100 (rank : 35) | θ value | 0.00102713 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.468057 (rank : 36) | θ value | 0.000121331 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.467267 (rank : 37) | θ value | 2.44474e-05 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
FKBP8_MOUSE
|
||||||
| NC score | 0.425244 (rank : 38) | θ value | 0.365318 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
PPP5_HUMAN
|
||||||
| NC score | 0.415796 (rank : 39) | θ value | 3.29651e-10 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.415546 (rank : 40) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
AIP_HUMAN
|
||||||
| NC score | 0.414462 (rank : 41) | θ value | 2.88788e-06 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
PPP5_MOUSE
|
||||||
| NC score | 0.414426 (rank : 42) | θ value | 4.30538e-10 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.413248 (rank : 43) | θ value | 5.44631e-05 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIP_MOUSE
|
||||||
| NC score | 0.403265 (rank : 44) | θ value | 1.43324e-05 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
AIPL1_MOUSE
|
||||||
| NC score | 0.403184 (rank : 45) | θ value | 0.00035302 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.391441 (rank : 46) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
Z3H7A_HUMAN
|
||||||
| NC score | 0.389092 (rank : 47) | θ value | 0.0148317 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
WDTC1_MOUSE
|
||||||
| NC score | 0.375066 (rank : 48) | θ value | 2.61198e-07 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_HUMAN
|
||||||
| NC score | 0.371534 (rank : 49) | θ value | 7.59969e-07 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.356358 (rank : 50) | θ value | 4.1701e-05 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
SUGT1_MOUSE
|
||||||
| NC score | 0.332534 (rank : 51) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
TTC13_HUMAN
|
||||||
| NC score | 0.319198 (rank : 52) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.317923 (rank : 53) | θ value | 0.00509761 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.315592 (rank : 54) | θ value | 0.00035302 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.310644 (rank : 55) | θ value | 0.0961366 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.304853 (rank : 56) | θ value | 0.0431538 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.302442 (rank : 57) | θ value | 0.00509761 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
BBS4_HUMAN
|
||||||
| NC score | 0.287030 (rank : 58) | θ value | 0.0252991 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| NC score | 0.282034 (rank : 59) | θ value | 0.0563607 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
K0372_HUMAN
|
||||||
| NC score | 0.273019 (rank : 60) | θ value | 0.0736092 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
SUGT1_HUMAN
|
||||||
| NC score | 0.269651 (rank : 61) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
TTC16_MOUSE
|
||||||
| NC score | 0.263329 (rank : 62) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.245875 (rank : 63) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.231513 (rank : 64) | θ value | 0.0330416 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
IFT88_HUMAN
|
||||||
| NC score | 0.225553 (rank : 65) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.224600 (rank : 66) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.224224 (rank : 67) | θ value | 0.0330416 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.212378 (rank : 68) | θ value | 0.163984 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.208004 (rank : 69) | θ value | 0.279714 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.206776 (rank : 70) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
FKBP6_HUMAN
|
||||||
| NC score | 0.200203 (rank : 71) | θ value | 0.0330416 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_MOUSE
|
||||||
| NC score | 0.194755 (rank : 72) | θ value | 0.0961366 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.130061 (rank : 73) | θ value | 0.813845 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.129152 (rank : 74) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
FKB1A_HUMAN
|
||||||
| NC score | 0.110332 (rank : 75) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1A_MOUSE
|
||||||
| NC score | 0.109491 (rank : 76) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1B_HUMAN
|
||||||
| NC score | 0.105328 (rank : 77) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
PEX5_MOUSE
|
||||||
| NC score | 0.105088 (rank : 78) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
FKB1B_MOUSE
|
||||||
| NC score | 0.104857 (rank : 79) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
GPSM2_MOUSE
|
||||||
| NC score | 0.102259 (rank : 80) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
PEX5_HUMAN
|
||||||
| NC score | 0.101828 (rank : 81) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
GPSM2_HUMAN
|
||||||
| NC score | 0.100569 (rank : 82) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM1_MOUSE
|
||||||
| NC score | 0.098623 (rank : 83) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.093273 (rank : 84) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.092407 (rank : 85) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
FKBP2_MOUSE
|
||||||
| NC score | 0.092038 (rank : 86) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_HUMAN
|
||||||
| NC score | 0.091520 (rank : 87) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
GPSM1_HUMAN
|
||||||
| NC score | 0.088751 (rank : 88) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86YR5, Q86SR5, Q969T1, Q9UFS8 | Gene names | GPSM1, AGS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
FKBP3_HUMAN
|
||||||
| NC score | 0.088272 (rank : 89) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
CDC23_HUMAN
|
||||||
| NC score | 0.087802 (rank : 90) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
FKBP3_MOUSE
|
||||||
| NC score | 0.087327 (rank : 91) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
CDC23_MOUSE
|
||||||
| NC score | 0.085398 (rank : 92) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
APC7_HUMAN
|
||||||
| NC score | 0.084625 (rank : 93) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
APC7_MOUSE
|
||||||
| NC score | 0.082999 (rank : 94) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
PEX5R_MOUSE
|
||||||
| NC score | 0.079913 (rank : 95) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
PEX5R_HUMAN
|
||||||
| NC score | 0.078631 (rank : 96) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
FKB11_MOUSE
|
||||||
| NC score | 0.071805 (rank : 97) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB11_HUMAN
|
||||||
| NC score | 0.069454 (rank : 98) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKBP7_MOUSE
|
||||||
| NC score | 0.066914 (rank : 99) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
TPM3_MOUSE
|
||||||
| NC score | 0.066469 (rank : 100) | θ value | 0.00509761 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
TPM3_HUMAN
|
||||||
| NC score | 0.065889 (rank : 101) | θ value | 0.00509761 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
TTC8_HUMAN
|
||||||
| NC score | 0.065455 (rank : 102) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
IFIT3_HUMAN
|
||||||
| NC score | 0.064392 (rank : 103) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14879, Q99634, Q9BSK7 | Gene names | IFIT3, IFI60, IFIT4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (IFIT-4) (Interferon-induced 60 kDa protein) (IFI-60K) (ISG-60) (CIG49) (Retinoic acid-induced gene G protein) (RIG-G). | |||||
|
RAPSN_HUMAN
|
||||||
| NC score | 0.062201 (rank : 104) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13702, Q9BTD9 | Gene names | RAPSN | |||
|
Domain Architecture |

|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
FKB10_HUMAN
|
||||||
| NC score | 0.061882 (rank : 105) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB10_MOUSE
|
||||||
| NC score | 0.061345 (rank : 106) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61576, Q8VHI1 | Gene names | Fkbp10, Fkbp-rs, Fkbp1-rs, Fkbp6, Fkbprp | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
TTC8_MOUSE
|
||||||
| NC score | 0.060950 (rank : 107) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
FKBP9_MOUSE
|
||||||
| NC score | 0.060823 (rank : 108) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z247, Q80ZZ6, Q8R386, Q9CVM0, Q9JHX5 | Gene names | Fkbp9, Fkbp60, Fkbp63 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP65RS). | |||||
|
FKBP9_HUMAN
|
||||||
| NC score | 0.059983 (rank : 109) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95302, Q3MIR7, Q6IN76, Q6P2N1, Q96EX5, Q96IJ9 | Gene names | FKBP9, FKBP60, FKBP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
CTNB1_MOUSE
|
||||||
| NC score | 0.057037 (rank : 110) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_HUMAN
|
||||||
| NC score | 0.057021 (rank : 111) | θ value | 0.0736092 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
RAPSN_MOUSE
|
||||||
| NC score | 0.056905 (rank : 112) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |

|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
TTC7A_MOUSE
|
||||||
| NC score | 0.056123 (rank : 113) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGB2, Q80XT5 | Gene names | Ttc7a, Ttc7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
FKB14_MOUSE
|
||||||
| NC score | 0.055416 (rank : 114) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59024 | Gene names | Fkbp14 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
TPM4_HUMAN
|
||||||
| NC score | 0.054664 (rank : 115) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P67936, P07226, Q15659, Q9BU85, Q9H8Q3 | Gene names | TPM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4) (TM30p1). | |||||
|
IFIT1_HUMAN
|
||||||
| NC score | 0.052731 (rank : 116) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09914, Q96QM5 | Gene names | IFIT1, G10P1, IFI56 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K). | |||||
|
FKB14_HUMAN
|
||||||
| NC score | 0.052148 (rank : 117) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NWM8 | Gene names | FKBP14, FKBP22 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (22 kDa FK506-binding protein) (FKBP-22). | |||||
|
IFIT1_MOUSE
|
||||||
| NC score | 0.050006 (rank : 118) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64282 | Gene names | Ifit1, Garg16, Ifi56 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K) (Glucocorticoid- attenuated response gene 16 protein) (GARG-16). | |||||
|
TPM1_MOUSE
|
||||||
| NC score | 0.048409 (rank : 119) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_HUMAN
|
||||||
| NC score | 0.048086 (rank : 120) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.046278 (rank : 121) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
TPM4_MOUSE
|
||||||
| NC score | 0.043885 (rank : 122) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.039305 (rank : 123) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
RBP22_HUMAN
|
||||||
| NC score | 0.038442 (rank : 124) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RGPD8_HUMAN
|
||||||
| NC score | 0.037728 (rank : 125) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
ZMY12_HUMAN
|
||||||
| NC score | 0.036945 (rank : 126) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.036587 (rank : 127) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
SIL1_MOUSE
|
||||||
| NC score | 0.029138 (rank : 128) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EPK6, Q91V34 | Gene names | Sil1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor. | |||||
|
SIL1_HUMAN
|
||||||
| NC score | 0.028643 (rank : 129) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H173, Q8N2L3 | Gene names | SIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor (BiP-associated protein) (BAP). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.026849 (rank : 130) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.026732 (rank : 131) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.026629 (rank : 132) | θ value | 0.365318 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.026583 (rank : 133) | θ value | 0.47712 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.025992 (rank : 134) | θ value | 0.47712 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.025709 (rank : 135) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.024436 (rank : 136) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
RPTOR_HUMAN
|
||||||
| NC score | 0.023659 (rank : 137) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N122, Q8N4V9, Q8TB32, Q9P2P3 | Gene names | RAPTOR, KIAA1303 | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.023476 (rank : 138) | θ value | 0.365318 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.023159 (rank : 139) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
INSC_HUMAN
|
||||||
| NC score | 0.022801 (rank : 140) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q1MX18, Q1MX19, Q3C1V6, Q4AC95, Q4AC96, Q4AC97, Q4AC98 | Gene names | INSC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inscuteable homolog. | |||||
|
RPTOR_MOUSE
|
||||||
| NC score | 0.022761 (rank : 141) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K4Q0, Q8C9W9, Q8CBY4, Q8CDY8, Q9D4H3 | Gene names | Raptor | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.022022 (rank : 142) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.021621 (rank : 143) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.021291 (rank : 144) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.020137 (rank : 145) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.019088 (rank : 146) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.018188 (rank : 147) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
TSH2_MOUSE
|
||||||
| NC score | 0.017708 (rank : 148) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
TSH1_HUMAN
|
||||||
| NC score | 0.017300 (rank : 149) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.017028 (rank : 150) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.016865 (rank : 151) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
TSH2_HUMAN
|
||||||
| NC score | 0.016174 (rank : 152) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.015993 (rank : 153) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.014934 (rank : 154) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.013309 (rank : 155) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.011870 (rank : 156) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
RB_HUMAN
|
||||||
| NC score | 0.011691 (rank : 157) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06400, P78499, Q5VW46, Q8IZL4 | Gene names | RB1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-associated protein (PP110) (P105-RB) (RB). | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.011131 (rank : 158) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
CP39A_HUMAN
|
||||||
| NC score | 0.009570 (rank : 159) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.008002 (rank : 160) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
HS70L_MOUSE
|
||||||
| NC score | 0.003373 (rank : 161) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||