Please be patient as the page loads
|
PPP5_MOUSE
|
||||||
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PPP5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999565 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
PPP5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
PP2AB_HUMAN
|
||||||
| θ value | 1.19377e-60 (rank : 3) | NC score | 0.855422 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62714, P11082 | Gene names | PPP2CB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform (EC 3.1.3.16) (PP2A-beta). | |||||
|
PP2AB_MOUSE
|
||||||
| θ value | 1.19377e-60 (rank : 4) | NC score | 0.855422 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62715, P11082 | Gene names | Ppp2cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform (EC 3.1.3.16) (PP2A-beta). | |||||
|
PPE1_HUMAN
|
||||||
| θ value | 7.73779e-60 (rank : 5) | NC score | 0.814382 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14829, O15253, Q9NU21, Q9UJH0 | Gene names | PPEF1, PPEF, PPP7C | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 1 (EC 3.1.3.16) (PPEF-1) (Protein phosphatase with EF calcium-binding domain) (PPEF) (Serine/threonine-protein phosphatase 7) (PP7). | |||||
|
PP2AA_HUMAN
|
||||||
| θ value | 1.7238e-59 (rank : 6) | NC score | 0.854451 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P67775, P05323, P13197 | Gene names | PPP2CA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform (EC 3.1.3.16) (PP2A-alpha) (Replication protein C) (RP-C). | |||||
|
PP2AA_MOUSE
|
||||||
| θ value | 1.7238e-59 (rank : 7) | NC score | 0.854456 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63330, O88591, P13353, Q5SNY5 | Gene names | Ppp2ca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform (EC 3.1.3.16) (PP2A-alpha). | |||||
|
PP4C_HUMAN
|
||||||
| θ value | 2.7521e-57 (rank : 8) | NC score | 0.855500 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60510, P33172 | Gene names | PPP4C, PPP4, PPX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 catalytic subunit (EC 3.1.3.16) (PP4C) (Pp4) (Protein phosphatase X) (PP-X). | |||||
|
PP4C_MOUSE
|
||||||
| θ value | 2.7521e-57 (rank : 9) | NC score | 0.855500 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97470, P33172 | Gene names | Ppp4c, Ppp4, Ppx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 catalytic subunit (EC 3.1.3.16) (PP4C) (Pp4) (Protein phosphatase X) (PP-X). | |||||
|
PP2BB_HUMAN
|
||||||
| θ value | 9.78833e-55 (rank : 10) | NC score | 0.831478 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P16298, P16299, Q8N3W4 | Gene names | PPP3CB, CALNA2, CNA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit beta isoform) (CAM-PRP catalytic subunit). | |||||
|
PP2BB_MOUSE
|
||||||
| θ value | 9.78833e-55 (rank : 11) | NC score | 0.831480 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48453, Q6NZR4 | Gene names | Ppp3cb, Calnb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit beta isoform) (CAM-PRP catalytic subunit). | |||||
|
PP1A_HUMAN
|
||||||
| θ value | 3.71957e-54 (rank : 12) | NC score | 0.849071 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62136, P08129, P20653, P22802 | Gene names | PPP1CA, PPP1A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit (EC 3.1.3.16) (PP-1A). | |||||
|
PP1A_MOUSE
|
||||||
| θ value | 3.71957e-54 (rank : 13) | NC score | 0.849073 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62137, P08129, P20653, P22802, Q3U7G7, Q9Z1G2 | Gene names | Ppp1ca, Ppp1a | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit (EC 3.1.3.16) (PP-1A). | |||||
|
PP1G_HUMAN
|
||||||
| θ value | 8.28633e-54 (rank : 14) | NC score | 0.849348 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P36873 | Gene names | PPP1CC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-gamma catalytic subunit (EC 3.1.3.16) (PP-1G) (Protein phosphatase 1C catalytic subunit). | |||||
|
PP1G_MOUSE
|
||||||
| θ value | 8.28633e-54 (rank : 15) | NC score | 0.849348 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63087, O09186, O09189, P37139, Q64679 | Gene names | Ppp1cc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase PP1-gamma catalytic subunit (EC 3.1.3.16) (PP-1G) (Protein phosphatase 1C catalytic subunit). | |||||
|
PP2BA_HUMAN
|
||||||
| θ value | 8.28633e-54 (rank : 16) | NC score | 0.832245 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q08209, Q8TAW9 | Gene names | PPP3CA, CALNA, CNA | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit alpha isoform) (CAM-PRP catalytic subunit). | |||||
|
PP2BA_MOUSE
|
||||||
| θ value | 1.08223e-53 (rank : 17) | NC score | 0.832088 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63328, P12816, P20652, Q3UCU1, Q64135 | Gene names | Ppp3ca, Calna | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit alpha isoform) (CAM-PRP catalytic subunit). | |||||
|
PP1B_HUMAN
|
||||||
| θ value | 3.1488e-53 (rank : 18) | NC score | 0.848905 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62140, P37140, Q5U087, Q6FG45 | Gene names | PPP1CB | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-beta catalytic subunit (EC 3.1.3.16) (PP-1B). | |||||
|
PP1B_MOUSE
|
||||||
| θ value | 3.1488e-53 (rank : 19) | NC score | 0.848905 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62141, P37140 | Gene names | Ppp1cb | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-beta catalytic subunit (EC 3.1.3.16) (PP-1B). | |||||
|
PP2BC_HUMAN
|
||||||
| θ value | 5.37103e-53 (rank : 20) | NC score | 0.832325 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48454 | Gene names | PPP3CC, CALNA3, CNA3 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit gamma isoform) (Calcineurin, testis-specific catalytic subunit) (CAM- PRP catalytic subunit). | |||||
|
PPP6_MOUSE
|
||||||
| θ value | 9.16162e-53 (rank : 21) | NC score | 0.853655 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CQR6 | Gene names | Ppp6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 6 (EC 3.1.3.16) (PP6). | |||||
|
PPP6_HUMAN
|
||||||
| θ value | 1.56274e-52 (rank : 22) | NC score | 0.853642 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00743 | Gene names | PPP6C, PPP6 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 6 (EC 3.1.3.16) (PP6). | |||||
|
PP2BC_MOUSE
|
||||||
| θ value | 5.02714e-51 (rank : 23) | NC score | 0.831135 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48455 | Gene names | Ppp3cc, Calnc | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit gamma isoform) (Calcineurin, testis-specific catalytic subunit) (CAM- PRP catalytic subunit). | |||||
|
PPE2_HUMAN
|
||||||
| θ value | 5.21438e-40 (rank : 24) | NC score | 0.769995 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14830, O14831 | Gene names | PPEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
PPE2_MOUSE
|
||||||
| θ value | 2.58786e-39 (rank : 25) | NC score | 0.766714 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 26) | NC score | 0.346224 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
SGTA_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 27) | NC score | 0.466411 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 28) | NC score | 0.343611 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
SGTB_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 29) | NC score | 0.479781 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |
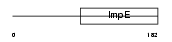
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
SGTB_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 30) | NC score | 0.480534 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
SGTA_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 31) | NC score | 0.464073 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |
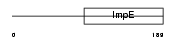
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
OM34_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 32) | NC score | 0.462221 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |
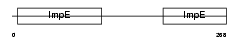
|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
OM34_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 33) | NC score | 0.455874 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 34) | NC score | 0.389731 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 35) | NC score | 0.434589 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 36) | NC score | 0.431480 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
TTC3_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 37) | NC score | 0.239636 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
UN45B_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 38) | NC score | 0.378188 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 39) | NC score | 0.418368 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
UN45B_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 40) | NC score | 0.380129 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 41) | NC score | 0.371103 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
TOM70_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 42) | NC score | 0.428499 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 43) | NC score | 0.429466 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 44) | NC score | 0.402679 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC1_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 45) | NC score | 0.444039 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 46) | NC score | 0.419396 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
TTC12_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 47) | NC score | 0.414426 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
STUB1_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 48) | NC score | 0.446734 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
STUB1_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 49) | NC score | 0.440683 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
WDTC1_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 50) | NC score | 0.249059 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
TTC12_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 51) | NC score | 0.406631 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |
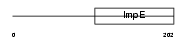
|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
WDTC1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 52) | NC score | 0.246842 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
FKBP4_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 53) | NC score | 0.277654 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 54) | NC score | 0.370353 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
FKBP4_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 55) | NC score | 0.273505 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 56) | NC score | 0.287434 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
F10A4_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 57) | NC score | 0.331095 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
TTC4_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 58) | NC score | 0.376801 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
F10A1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 59) | NC score | 0.309470 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
FKBP5_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 60) | NC score | 0.276429 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
F10A5_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 61) | NC score | 0.312621 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
F10A1_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 62) | NC score | 0.327209 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
PPID_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 63) | NC score | 0.187023 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
DYXC1_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 64) | NC score | 0.376498 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
PPID_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 65) | NC score | 0.197795 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
TTC4_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 66) | NC score | 0.357591 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
Z3H7B_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 67) | NC score | 0.299128 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
K0372_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 68) | NC score | 0.198433 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 69) | NC score | 0.219534 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
TTC6_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 70) | NC score | 0.292451 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
SUGT1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 71) | NC score | 0.244498 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 72) | NC score | 0.192757 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
Z3H7A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 73) | NC score | 0.232032 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
TTC13_HUMAN
|
||||||
| θ value | 0.163984 (rank : 74) | NC score | 0.203162 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
BBS4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 75) | NC score | 0.168600 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 76) | NC score | 0.166862 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
CDC27_HUMAN
|
||||||
| θ value | 0.279714 (rank : 77) | NC score | 0.161844 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 78) | NC score | 0.187610 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | 0.365318 (rank : 79) | NC score | 0.167396 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
SUGT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 80) | NC score | 0.204365 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |
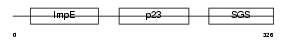
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 81) | NC score | 0.014489 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 82) | NC score | 0.014486 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 0.813845 (rank : 83) | NC score | 0.131450 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TTC16_MOUSE
|
||||||
| θ value | 0.813845 (rank : 84) | NC score | 0.178065 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
UTP11_MOUSE
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.036974 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CZJ1, Q3UI21, Q9CVB5 | Gene names | Utp11l | |||
|
Domain Architecture |
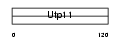
|
|||||
| Description | Probable U3 small nucleolar RNA-associated protein 11 (U3 snoRNA- associated protein 11) (UTP11-like protein). | |||||
|
TACC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.011670 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.073206 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.072748 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
PEX5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.081232 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
PEX5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.082876 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.009911 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.009165 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.009759 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
IFT88_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.099356 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.093211 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.000296 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
CRNL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.010840 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P63154, Q542E8, Q9CQC1 | Gene names | Crnkl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crooked neck-like protein 1 (Crooked neck homolog). | |||||
|
KIF1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.000886 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12756 | Gene names | KIF1A, ATSV | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
KIF1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.000895 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P33173, Q61770 | Gene names | Kif1a, Atsv, Kif1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
TTC17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.031156 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96AE7 | Gene names | TTC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 17 (TPR repeat protein 17). | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.140186 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.134985 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.130322 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
AIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.124845 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
APC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.055822 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
APC7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.054688 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
FKBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.068864 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |
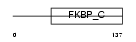
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.071654 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |
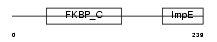
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.208073 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
FKBP8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.218220 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
IFT88_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.099370 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
PEX5R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.057158 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
PEX5R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.058551 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.093355 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.091278 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
PPP5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
PPP5_HUMAN
|
||||||
| NC score | 0.999565 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
PP4C_HUMAN
|
||||||
| NC score | 0.855500 (rank : 3) | θ value | 2.7521e-57 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60510, P33172 | Gene names | PPP4C, PPP4, PPX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 catalytic subunit (EC 3.1.3.16) (PP4C) (Pp4) (Protein phosphatase X) (PP-X). | |||||
|
PP4C_MOUSE
|
||||||
| NC score | 0.855500 (rank : 4) | θ value | 2.7521e-57 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97470, P33172 | Gene names | Ppp4c, Ppp4, Ppx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 catalytic subunit (EC 3.1.3.16) (PP4C) (Pp4) (Protein phosphatase X) (PP-X). | |||||
|
PP2AB_HUMAN
|
||||||
| NC score | 0.855422 (rank : 5) | θ value | 1.19377e-60 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62714, P11082 | Gene names | PPP2CB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform (EC 3.1.3.16) (PP2A-beta). | |||||
|
PP2AB_MOUSE
|
||||||
| NC score | 0.855422 (rank : 6) | θ value | 1.19377e-60 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62715, P11082 | Gene names | Ppp2cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform (EC 3.1.3.16) (PP2A-beta). | |||||
|
PP2AA_MOUSE
|
||||||
| NC score | 0.854456 (rank : 7) | θ value | 1.7238e-59 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63330, O88591, P13353, Q5SNY5 | Gene names | Ppp2ca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform (EC 3.1.3.16) (PP2A-alpha). | |||||
|
PP2AA_HUMAN
|
||||||
| NC score | 0.854451 (rank : 8) | θ value | 1.7238e-59 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P67775, P05323, P13197 | Gene names | PPP2CA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform (EC 3.1.3.16) (PP2A-alpha) (Replication protein C) (RP-C). | |||||
|
PPP6_MOUSE
|
||||||
| NC score | 0.853655 (rank : 9) | θ value | 9.16162e-53 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CQR6 | Gene names | Ppp6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 6 (EC 3.1.3.16) (PP6). | |||||
|
PPP6_HUMAN
|
||||||
| NC score | 0.853642 (rank : 10) | θ value | 1.56274e-52 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00743 | Gene names | PPP6C, PPP6 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 6 (EC 3.1.3.16) (PP6). | |||||
|
PP1G_HUMAN
|
||||||
| NC score | 0.849348 (rank : 11) | θ value | 8.28633e-54 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P36873 | Gene names | PPP1CC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-gamma catalytic subunit (EC 3.1.3.16) (PP-1G) (Protein phosphatase 1C catalytic subunit). | |||||
|
PP1G_MOUSE
|
||||||
| NC score | 0.849348 (rank : 12) | θ value | 8.28633e-54 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63087, O09186, O09189, P37139, Q64679 | Gene names | Ppp1cc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase PP1-gamma catalytic subunit (EC 3.1.3.16) (PP-1G) (Protein phosphatase 1C catalytic subunit). | |||||
|
PP1A_MOUSE
|
||||||
| NC score | 0.849073 (rank : 13) | θ value | 3.71957e-54 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62137, P08129, P20653, P22802, Q3U7G7, Q9Z1G2 | Gene names | Ppp1ca, Ppp1a | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit (EC 3.1.3.16) (PP-1A). | |||||
|
PP1A_HUMAN
|
||||||
| NC score | 0.849071 (rank : 14) | θ value | 3.71957e-54 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62136, P08129, P20653, P22802 | Gene names | PPP1CA, PPP1A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit (EC 3.1.3.16) (PP-1A). | |||||
|
PP1B_HUMAN
|
||||||
| NC score | 0.848905 (rank : 15) | θ value | 3.1488e-53 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62140, P37140, Q5U087, Q6FG45 | Gene names | PPP1CB | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-beta catalytic subunit (EC 3.1.3.16) (PP-1B). | |||||
|
PP1B_MOUSE
|
||||||
| NC score | 0.848905 (rank : 16) | θ value | 3.1488e-53 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62141, P37140 | Gene names | Ppp1cb | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase PP1-beta catalytic subunit (EC 3.1.3.16) (PP-1B). | |||||
|
PP2BC_HUMAN
|
||||||
| NC score | 0.832325 (rank : 17) | θ value | 5.37103e-53 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48454 | Gene names | PPP3CC, CALNA3, CNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit gamma isoform) (Calcineurin, testis-specific catalytic subunit) (CAM- PRP catalytic subunit). | |||||
|
PP2BA_HUMAN
|
||||||
| NC score | 0.832245 (rank : 18) | θ value | 8.28633e-54 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q08209, Q8TAW9 | Gene names | PPP3CA, CALNA, CNA | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit alpha isoform) (CAM-PRP catalytic subunit). | |||||
|
PP2BA_MOUSE
|
||||||
| NC score | 0.832088 (rank : 19) | θ value | 1.08223e-53 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63328, P12816, P20652, Q3UCU1, Q64135 | Gene names | Ppp3ca, Calna | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit alpha isoform) (CAM-PRP catalytic subunit). | |||||
|
PP2BB_MOUSE
|
||||||
| NC score | 0.831480 (rank : 20) | θ value | 9.78833e-55 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48453, Q6NZR4 | Gene names | Ppp3cb, Calnb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit beta isoform) (CAM-PRP catalytic subunit). | |||||
|
PP2BB_HUMAN
|
||||||
| NC score | 0.831478 (rank : 21) | θ value | 9.78833e-55 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P16298, P16299, Q8N3W4 | Gene names | PPP3CB, CALNA2, CNA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit beta isoform) (CAM-PRP catalytic subunit). | |||||
|
PP2BC_MOUSE
|
||||||
| NC score | 0.831135 (rank : 22) | θ value | 5.02714e-51 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48455 | Gene names | Ppp3cc, Calnc | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit gamma isoform) (Calcineurin, testis-specific catalytic subunit) (CAM- PRP catalytic subunit). | |||||
|
PPE1_HUMAN
|
||||||
| NC score | 0.814382 (rank : 23) | θ value | 7.73779e-60 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14829, O15253, Q9NU21, Q9UJH0 | Gene names | PPEF1, PPEF, PPP7C | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 1 (EC 3.1.3.16) (PPEF-1) (Protein phosphatase with EF calcium-binding domain) (PPEF) (Serine/threonine-protein phosphatase 7) (PP7). | |||||
|
PPE2_HUMAN
|
||||||
| NC score | 0.769995 (rank : 24) | θ value | 5.21438e-40 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14830, O14831 | Gene names | PPEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
PPE2_MOUSE
|
||||||
| NC score | 0.766714 (rank : 25) | θ value | 2.58786e-39 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
SGTB_MOUSE
|
||||||
| NC score | 0.480534 (rank : 26) | θ value | 4.02038e-16 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
SGTB_HUMAN
|
||||||
| NC score | 0.479781 (rank : 27) | θ value | 4.02038e-16 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
SGTA_HUMAN
|
||||||
| NC score | 0.466411 (rank : 28) | θ value | 4.74913e-17 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
SGTA_MOUSE
|
||||||
| NC score | 0.464073 (rank : 29) | θ value | 8.95645e-16 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
OM34_MOUSE
|
||||||
| NC score | 0.462221 (rank : 30) | θ value | 1.68911e-14 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
OM34_HUMAN
|
||||||
| NC score | 0.455874 (rank : 31) | θ value | 4.91457e-14 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.446734 (rank : 32) | θ value | 7.34386e-10 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.444039 (rank : 33) | θ value | 1.9326e-10 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.440683 (rank : 34) | θ value | 2.79066e-09 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.434589 (rank : 35) | θ value | 1.86753e-13 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.431480 (rank : 36) | θ value | 7.09661e-13 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
TOM70_MOUSE
|
||||||
| NC score | 0.429466 (rank : 37) | θ value | 1.133e-10 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.428499 (rank : 38) | θ value | 2.28291e-11 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.419396 (rank : 39) | θ value | 2.52405e-10 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.418368 (rank : 40) | θ value | 6.00763e-12 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.414426 (rank : 41) | θ value | 4.30538e-10 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC12_MOUSE
|
||||||
| NC score | 0.406631 (rank : 42) | θ value | 1.38499e-08 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.402679 (rank : 43) | θ value | 1.9326e-10 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.389731 (rank : 44) | θ value | 1.42992e-13 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
UN45B_MOUSE
|
||||||
| NC score | 0.380129 (rank : 45) | θ value | 6.00763e-12 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
UN45B_HUMAN
|
||||||
| NC score | 0.378188 (rank : 46) | θ value | 3.52202e-12 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
TTC4_HUMAN
|
||||||
| NC score | 0.376801 (rank : 47) | θ value | 1.69304e-06 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
DYXC1_MOUSE
|
||||||
| NC score | 0.376498 (rank : 48) | θ value | 2.44474e-05 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.371103 (rank : 49) | θ value | 1.74796e-11 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.370353 (rank : 50) | θ value | 1.53129e-07 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
TTC4_MOUSE
|
||||||
| NC score | 0.357591 (rank : 51) | θ value | 3.19293e-05 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.346224 (rank : 52) | θ value | 2.7842e-17 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.343611 (rank : 53) | θ value | 8.10077e-17 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.331095 (rank : 54) | θ value | 9.92553e-07 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
F10A1_MOUSE
|
||||||
| NC score | 0.327209 (rank : 55) | θ value | 6.43352e-06 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.312621 (rank : 56) | θ value | 4.92598e-06 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.309470 (rank : 57) | θ value | 2.21117e-06 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
Z3H7B_HUMAN
|
||||||
| NC score | 0.299128 (rank : 58) | θ value | 5.44631e-05 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.292451 (rank : 59) | θ value | 0.00102713 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
FKBP5_HUMAN
|
||||||
| NC score | 0.287434 (rank : 60) | θ value | 2.61198e-07 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP4_HUMAN
|
||||||
| NC score | 0.277654 (rank : 61) | θ value | 5.26297e-08 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_MOUSE
|
||||||
| NC score | 0.276429 (rank : 62) | θ value | 3.77169e-06 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
FKBP4_MOUSE
|
||||||
| NC score | 0.273505 (rank : 63) | θ value | 1.99992e-07 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
WDTC1_MOUSE
|
||||||
| NC score | 0.249059 (rank : 64) | θ value | 8.11959e-09 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_HUMAN
|
||||||
| NC score | 0.246842 (rank : 65) | θ value | 2.36244e-08 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
SUGT1_MOUSE
|
||||||
| NC score | 0.244498 (rank : 66) | θ value | 0.0736092 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.239636 (rank : 67) | θ value | 3.52202e-12 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
Z3H7A_HUMAN
|
||||||
| NC score | 0.232032 (rank : 68) | θ value | 0.125558 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.219534 (rank : 69) | θ value | 0.000461057 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
FKBP8_MOUSE
|
||||||
| NC score | 0.218220 (rank : 70) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.208073 (rank : 71) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
SUGT1_HUMAN
|
||||||
| NC score | 0.204365 (rank : 72) | θ value | 0.62314 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
TTC13_HUMAN
|
||||||
| NC score | 0.203162 (rank : 73) | θ value | 0.163984 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
K0372_HUMAN
|
||||||
| NC score | 0.198433 (rank : 74) | θ value | 7.1131e-05 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.197795 (rank : 75) | θ value | 2.44474e-05 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.192757 (rank : 76) | θ value | 0.125558 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.187610 (rank : 77) | θ value | 0.279714 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.187023 (rank : 78) | θ value | 1.87187e-05 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
TTC16_MOUSE
|
||||||
| NC score | 0.178065 (rank : 79) | θ value | 0.813845 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
BBS4_HUMAN
|
||||||
| NC score | 0.168600 (rank : 80) | θ value | 0.21417 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.167396 (rank : 81) | θ value | 0.365318 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
BBS4_MOUSE
|
||||||
| NC score | 0.166862 (rank : 82) | θ value | 0.21417 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.161844 (rank : 83) | θ value | 0.279714 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.140186 (rank : 84) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_MOUSE
|
||||||
| NC score | 0.134985 (rank : 85) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.131450 (rank : 86) | θ value | 0.813845 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
AIP_HUMAN
|
||||||
| NC score | 0.130322 (rank : 87) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
AIP_MOUSE
|
||||||
| NC score | 0.124845 (rank : 88) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
IFT88_HUMAN
|
||||||
| NC score | 0.099370 (rank : 89) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.099356 (rank : 90) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.093355 (rank : 91) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.093211 (rank : 92) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.091278 (rank : 93) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
PEX5_MOUSE
|
||||||
| NC score | 0.082876 (rank : 94) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
PEX5_HUMAN
|
||||||
| NC score | 0.081232 (rank : 95) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.073206 (rank : 96) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.072748 (rank : 97) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
FKBP6_MOUSE
|
||||||
| NC score | 0.071654 (rank : 98) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_HUMAN
|
||||||
| NC score | 0.068864 (rank : 99) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
PEX5R_MOUSE
|
||||||
| NC score | 0.058551 (rank : 100) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
PEX5R_HUMAN
|
||||||
| NC score | 0.057158 (rank : 101) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
APC7_HUMAN
|
||||||
| NC score | 0.055822 (rank : 102) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
APC7_MOUSE
|
||||||
| NC score | 0.054688 (rank : 103) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
UTP11_MOUSE
|
||||||
| NC score | 0.036974 (rank : 104) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CZJ1, Q3UI21, Q9CVB5 | Gene names | Utp11l | |||
|
Domain Architecture |

|
|||||
| Description | Probable U3 small nucleolar RNA-associated protein 11 (U3 snoRNA- associated protein 11) (UTP11-like protein). | |||||
|
TTC17_HUMAN
|
||||||
| NC score | 0.031156 (rank : 105) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96AE7 | Gene names | TTC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 17 (TPR repeat protein 17). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.014489 (rank : 106) | θ value | 0.62314 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.014486 (rank : 107) | θ value | 0.62314 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
TACC1_HUMAN
|
||||||
| NC score | 0.011670 (rank : 108) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
CRNL1_MOUSE
|
||||||
| NC score | 0.010840 (rank : 109) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P63154, Q542E8, Q9CQC1 | Gene names | Crnkl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crooked neck-like protein 1 (Crooked neck homolog). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.009911 (rank : 110) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.009759 (rank : 111) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.009165 (rank : 112) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
KIF1A_MOUSE
|
||||||
| NC score | 0.000895 (rank : 113) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P33173, Q61770 | Gene names | Kif1a, Atsv, Kif1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
KIF1A_HUMAN
|
||||||
| NC score | 0.000886 (rank : 114) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12756 | Gene names | KIF1A, ATSV | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.000296 (rank : 115) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||