Please be patient as the page loads
|
SRTD2_HUMAN
|
||||||
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SRTD2_HUMAN
|
||||||
| θ value | 1.51632e-156 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
SRTD2_MOUSE
|
||||||
| θ value | 3.29472e-127 (rank : 2) | NC score | 0.962784 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJG5, Q8C609, Q91WL3, Q925E5 | Gene names | Sertad2, Kiaa0127 | |||
|
Domain Architecture |
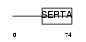
|
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
CDCA4_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 3) | NC score | 0.580186 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CWM2, Q3U1C0, Q921E8, Q99MP6 | Gene names | Cdca4, Hepp | |||
|
Domain Architecture |
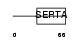
|
|||||
| Description | Cell division cycle-associated protein 4 (Hematopoietic progenitor protein). | |||||
|
CDCA4_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 4) | NC score | 0.572487 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXL8, Q8TB18, Q9NWK7 | Gene names | CDCA4, HEPP | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle-associated protein 4 (Hematopoietic progenitor protein). | |||||
|
SRTD1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 5) | NC score | 0.440015 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JL10, Q925E6, Q9D888 | Gene names | Sertad1, Sei1 | |||
|
Domain Architecture |
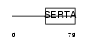
|
|||||
| Description | SERTA domain-containing protein 1 (Transcriptional regulator interacting with the PHD-bromodomain 1) (TRIP-Br1) (CDK4-binding protein p34SEI1) (SEI-1). | |||||
|
SRTD1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 6) | NC score | 0.450474 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHV2, Q9BUE7 | Gene names | SERTAD1, SEI1 | |||
|
Domain Architecture |
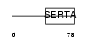
|
|||||
| Description | SERTA domain-containing protein 1 (Transcriptional regulator interacting with the PHD-bromodomain 1) (TRIP-Br1) (CDK4-binding protein p34SEI1) (SEI-1). | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.070793 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.070510 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.052190 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
VASN_MOUSE
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.035817 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CZT5, Q8BJJ0, Q8R2G5 | Gene names | Vasn, Slitl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.045202 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
VASN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.033130 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6EMK4, Q6UXL4, Q6UXL5, Q96CX1 | Gene names | VASN, SLITL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.062348 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.024368 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.072760 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.045594 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.064000 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.082313 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.046081 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CU056_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.053083 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0A9, Q9NSE5 | Gene names | C21orf56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56. | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.042446 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.014358 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.036836 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
NR1D1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.015233 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.042841 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.068869 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.043815 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
VINEX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.022211 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60504, Q9UQE4 | Gene names | SORBS3, SCAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.013568 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
PARM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.075676 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.020606 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.025184 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
IER5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.035514 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
PTN12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.013269 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
KCC2G_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.001420 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13555, O15378, Q13556, Q5SQZ4 | Gene names | CAMK2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II gamma chain (EC 2.7.11.17) (CaM-kinase II gamma chain) (CaM kinase II gamma subunit) (CaMK-II subunit gamma). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.027746 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NPCL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.018877 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6T3U4, Q5SVX1 | Gene names | Npc1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niemann-Pick C1-like protein 1 precursor. | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.033448 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SULF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.016633 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFG0, Q8BM68, Q8BUZ4, Q8BX28, Q8BZ51, Q8C169, Q9D8E2 | Gene names | Sulf2 | |||
|
Domain Architecture |
|
|||||
| Description | Extracellular sulfatase Sulf-2 precursor (EC 3.1.6.-) (MSulf-2). | |||||
|
AAKG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.016948 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.030085 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.006098 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MUG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.012317 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28665 | Gene names | Mug1, Mug-1 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-1 precursor (MuG1). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.007770 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.032652 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
TLE6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.014095 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVB3 | Gene names | Tle6, Grg6 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 6 (Groucho-related protein 6). | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.014231 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
CN037_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.025426 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
LIMK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.000699 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53668 | Gene names | Limk1, Limk | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain kinase 1 (EC 2.7.11.1) (LIMK-1) (KIZ-1). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.023121 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.017194 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
CBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.012735 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.008853 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
KSR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.002256 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.013119 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PCTK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.002251 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04735 | Gene names | Pctk1, Crk5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PCTAIRE-1 (EC 2.7.11.22) (PCTAIRE- motif protein kinase 1) (CRK5). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.011857 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PRR8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.024436 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
SRTD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.098211 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJW9, Q96CQ2 | Gene names | SERTAD3, RBT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 3 (Replication protein-binding trans- activator) (RPA-binding trans-activator). | |||||
|
TLR7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.012844 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYK1, Q9NR98 | Gene names | TLR7 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 7 precursor. | |||||
|
TMM24_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.014497 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.009522 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
SRTD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.060677 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERC3 | Gene names | Sertad3, Rbt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 3 (Replication protein-binding trans- activator) (RPA-binding trans-activator). | |||||
|
SRTD2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.51632e-156 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
SRTD2_MOUSE
|
||||||
| NC score | 0.962784 (rank : 2) | θ value | 3.29472e-127 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJG5, Q8C609, Q91WL3, Q925E5 | Gene names | Sertad2, Kiaa0127 | |||
|
Domain Architecture |

|
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
CDCA4_MOUSE
|
||||||
| NC score | 0.580186 (rank : 3) | θ value | 1.133e-10 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CWM2, Q3U1C0, Q921E8, Q99MP6 | Gene names | Cdca4, Hepp | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle-associated protein 4 (Hematopoietic progenitor protein). | |||||
|
CDCA4_HUMAN
|
||||||
| NC score | 0.572487 (rank : 4) | θ value | 2.13673e-09 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXL8, Q8TB18, Q9NWK7 | Gene names | CDCA4, HEPP | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle-associated protein 4 (Hematopoietic progenitor protein). | |||||
|
SRTD1_HUMAN
|
||||||
| NC score | 0.450474 (rank : 5) | θ value | 8.40245e-06 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHV2, Q9BUE7 | Gene names | SERTAD1, SEI1 | |||
|
Domain Architecture |

|
|||||
| Description | SERTA domain-containing protein 1 (Transcriptional regulator interacting with the PHD-bromodomain 1) (TRIP-Br1) (CDK4-binding protein p34SEI1) (SEI-1). | |||||
|
SRTD1_MOUSE
|
||||||
| NC score | 0.440015 (rank : 6) | θ value | 1.69304e-06 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JL10, Q925E6, Q9D888 | Gene names | Sertad1, Sei1 | |||
|
Domain Architecture |

|
|||||
| Description | SERTA domain-containing protein 1 (Transcriptional regulator interacting with the PHD-bromodomain 1) (TRIP-Br1) (CDK4-binding protein p34SEI1) (SEI-1). | |||||
|
SRTD3_HUMAN
|
||||||
| NC score | 0.098211 (rank : 7) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJW9, Q96CQ2 | Gene names | SERTAD3, RBT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 3 (Replication protein-binding trans- activator) (RPA-binding trans-activator). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.082313 (rank : 8) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PARM1_HUMAN
|
||||||
| NC score | 0.075676 (rank : 9) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.072760 (rank : 10) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.070793 (rank : 11) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.070510 (rank : 12) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.068869 (rank : 13) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.064000 (rank : 14) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.062348 (rank : 15) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
SRTD3_MOUSE
|
||||||
| NC score | 0.060677 (rank : 16) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERC3 | Gene names | Sertad3, Rbt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 3 (Replication protein-binding trans- activator) (RPA-binding trans-activator). | |||||
|
CU056_HUMAN
|
||||||
| NC score | 0.053083 (rank : 17) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0A9, Q9NSE5 | Gene names | C21orf56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.052190 (rank : 18) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.046081 (rank : 19) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.045594 (rank : 20) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.045202 (rank : 21) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.043815 (rank : 22) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.042841 (rank : 23) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.042446 (rank : 24) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.036836 (rank : 25) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
VASN_MOUSE
|
||||||
| NC score | 0.035817 (rank : 26) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CZT5, Q8BJJ0, Q8R2G5 | Gene names | Vasn, Slitl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
IER5_MOUSE
|
||||||
| NC score | 0.035514 (rank : 27) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.033448 (rank : 28) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
VASN_HUMAN
|
||||||
| NC score | 0.033130 (rank : 29) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6EMK4, Q6UXL4, Q6UXL5, Q96CX1 | Gene names | VASN, SLITL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.032652 (rank : 30) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.030085 (rank : 31) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.027746 (rank : 32) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.025426 (rank : 33) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.025184 (rank : 34) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
PRR8_HUMAN
|
||||||
| NC score | 0.024436 (rank : 35) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.024368 (rank : 36) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.023121 (rank : 37) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
VINEX_HUMAN
|
||||||
| NC score | 0.022211 (rank : 38) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60504, Q9UQE4 | Gene names | SORBS3, SCAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.020606 (rank : 39) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
NPCL1_MOUSE
|
||||||
| NC score | 0.018877 (rank : 40) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6T3U4, Q5SVX1 | Gene names | Npc1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niemann-Pick C1-like protein 1 precursor. | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.017194 (rank : 41) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
AAKG2_HUMAN
|
||||||
| NC score | 0.016948 (rank : 42) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
SULF2_MOUSE
|
||||||
| NC score | 0.016633 (rank : 43) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFG0, Q8BM68, Q8BUZ4, Q8BX28, Q8BZ51, Q8C169, Q9D8E2 | Gene names | Sulf2 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-2 precursor (EC 3.1.6.-) (MSulf-2). | |||||
|
NR1D1_HUMAN
|
||||||
| NC score | 0.015233 (rank : 44) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
TMM24_MOUSE
|
||||||
| NC score | 0.014497 (rank : 45) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.014358 (rank : 46) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.014231 (rank : 47) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
TLE6_MOUSE
|
||||||
| NC score | 0.014095 (rank : 48) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVB3 | Gene names | Tle6, Grg6 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 6 (Groucho-related protein 6). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.013568 (rank : 49) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.013269 (rank : 50) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.013119 (rank : 51) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
TLR7_HUMAN
|
||||||
| NC score | 0.012844 (rank : 52) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYK1, Q9NR98 | Gene names | TLR7 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 7 precursor. | |||||
|
CBL_MOUSE
|
||||||
| NC score | 0.012735 (rank : 53) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
MUG1_MOUSE
|
||||||
| NC score | 0.012317 (rank : 54) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28665 | Gene names | Mug1, Mug-1 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-1 precursor (MuG1). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.011857 (rank : 55) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.009522 (rank : 56) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.008853 (rank : 57) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.007770 (rank : 58) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.006098 (rank : 59) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
KSR1_HUMAN
|
||||||
| NC score | 0.002256 (rank : 60) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
PCTK1_MOUSE
|
||||||
| NC score | 0.002251 (rank : 61) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04735 | Gene names | Pctk1, Crk5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PCTAIRE-1 (EC 2.7.11.22) (PCTAIRE- motif protein kinase 1) (CRK5). | |||||
|
KCC2G_HUMAN
|
||||||
| NC score | 0.001420 (rank : 62) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13555, O15378, Q13556, Q5SQZ4 | Gene names | CAMK2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II gamma chain (EC 2.7.11.17) (CaM-kinase II gamma chain) (CaM kinase II gamma subunit) (CaMK-II subunit gamma). | |||||
|
LIMK1_MOUSE
|
||||||
| NC score | 0.000699 (rank : 63) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53668 | Gene names | Limk1, Limk | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain kinase 1 (EC 2.7.11.1) (LIMK-1) (KIZ-1). | |||||