Please be patient as the page loads
|
CCNB1_MOUSE
|
||||||
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |
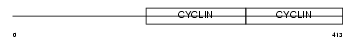
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCNB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995640 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |
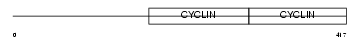
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_HUMAN
|
||||||
| θ value | 2.71407e-105 (rank : 3) | NC score | 0.983037 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |
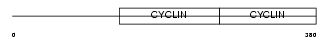
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB2_MOUSE
|
||||||
| θ value | 2.15048e-102 (rank : 4) | NC score | 0.982154 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |
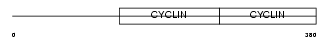
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNA1_HUMAN
|
||||||
| θ value | 2.67181e-44 (rank : 5) | NC score | 0.917645 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| θ value | 3.48949e-44 (rank : 6) | NC score | 0.917405 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |
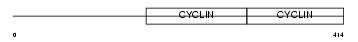
|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA2_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 7) | NC score | 0.907365 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 8) | NC score | 0.840566 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 1.3722e-40 (rank : 9) | NC score | 0.747427 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNA2_HUMAN
|
||||||
| θ value | 1.51715e-39 (rank : 10) | NC score | 0.905285 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNE2_MOUSE
|
||||||
| θ value | 1.2077e-20 (rank : 11) | NC score | 0.792233 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |
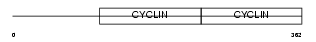
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE2_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 12) | NC score | 0.786608 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |
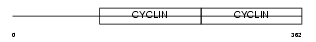
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 13) | NC score | 0.787218 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |
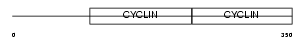
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE1_MOUSE
|
||||||
| θ value | 7.32683e-18 (rank : 14) | NC score | 0.778067 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNF_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 15) | NC score | 0.705307 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCND1_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 16) | NC score | 0.735961 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCNF_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 17) | NC score | 0.701730 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCND1_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 18) | NC score | 0.727666 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCND3_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 19) | NC score | 0.713870 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND3_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 20) | NC score | 0.702503 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |
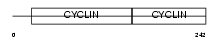
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND2_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 21) | NC score | 0.684794 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |
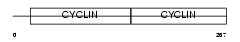
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND2_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 22) | NC score | 0.692094 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |
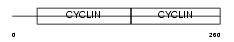
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCNI_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 23) | NC score | 0.484141 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |
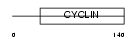
|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 24) | NC score | 0.506582 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNG1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 25) | NC score | 0.461532 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 26) | NC score | 0.474128 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.430817 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.433186 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
UNG2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.353582 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
NHRF2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.022749 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
IER2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.029588 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTL4, Q03827, Q2TAZ2 | Gene names | IER2, ETR101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 2 protein (Protein ETR101). | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.072138 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.072003 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.078447 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
DOCK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.010229 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14185 | Gene names | DOCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180). | |||||
|
CCNL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.068863 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
M3K4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | -0.000144 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
LMNB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.004623 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
TLR7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.003381 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYK1, Q9NR98 | Gene names | TLR7 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 7 precursor. | |||||
|
BUB1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.012350 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
SAFB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.005299 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.005842 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
GGPPS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.016657 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTN0 | Gene names | Ggps1 | |||
|
Domain Architecture |
|
|||||
| Description | Geranylgeranyl pyrophosphate synthetase (GGPP synthetase) (GGPPSase) (Geranylgeranyl diphosphate synthase) [Includes: Dimethylallyltranstransferase (EC 2.5.1.1); Geranyltranstransferase (EC 2.5.1.10); Farnesyltranstransferase (EC 2.5.1.29)]. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.022171 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.003070 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.000812 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
ZBTB5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | -0.002615 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 801 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQG0, Q3TVR7, Q6GV09, Q8BIB5 | Gene names | Zbtb5, Kiaa0354 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 5 (Transcription factor ZNF-POZ). | |||||
|
ZWINT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.006218 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
CCNK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.103151 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.084229 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNB1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_HUMAN
|
||||||
| NC score | 0.995640 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_HUMAN
|
||||||
| NC score | 0.983037 (rank : 3) | θ value | 2.71407e-105 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB2_MOUSE
|
||||||
| NC score | 0.982154 (rank : 4) | θ value | 2.15048e-102 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNA1_HUMAN
|
||||||
| NC score | 0.917645 (rank : 5) | θ value | 2.67181e-44 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| NC score | 0.917405 (rank : 6) | θ value | 3.48949e-44 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA2_MOUSE
|
||||||
| NC score | 0.907365 (rank : 7) | θ value | 1.05066e-40 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.905285 (rank : 8) | θ value | 1.51715e-39 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.840566 (rank : 9) | θ value | 1.05066e-40 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNE2_MOUSE
|
||||||
| NC score | 0.792233 (rank : 10) | θ value | 1.2077e-20 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_HUMAN
|
||||||
| NC score | 0.787218 (rank : 11) | θ value | 6.62687e-19 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE2_HUMAN
|
||||||
| NC score | 0.786608 (rank : 12) | θ value | 5.07402e-19 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_MOUSE
|
||||||
| NC score | 0.778067 (rank : 13) | θ value | 7.32683e-18 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.747427 (rank : 14) | θ value | 1.3722e-40 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCND1_MOUSE
|
||||||
| NC score | 0.735961 (rank : 15) | θ value | 1.52774e-15 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCND1_HUMAN
|
||||||
| NC score | 0.727666 (rank : 16) | θ value | 2.88119e-14 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCND3_HUMAN
|
||||||
| NC score | 0.713870 (rank : 17) | θ value | 7.09661e-13 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCNF_HUMAN
|
||||||
| NC score | 0.705307 (rank : 18) | θ value | 2.7842e-17 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCND3_MOUSE
|
||||||
| NC score | 0.702503 (rank : 19) | θ value | 1.02475e-11 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCNF_MOUSE
|
||||||
| NC score | 0.701730 (rank : 20) | θ value | 3.40345e-15 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCND2_HUMAN
|
||||||
| NC score | 0.692094 (rank : 21) | θ value | 8.67504e-11 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND2_MOUSE
|
||||||
| NC score | 0.684794 (rank : 22) | θ value | 6.64225e-11 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCNI_HUMAN
|
||||||
| NC score | 0.506582 (rank : 23) | θ value | 1.43324e-05 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_MOUSE
|
||||||
| NC score | 0.484141 (rank : 24) | θ value | 8.40245e-06 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNG1_MOUSE
|
||||||
| NC score | 0.474128 (rank : 25) | θ value | 0.00175202 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_HUMAN
|
||||||
| NC score | 0.461532 (rank : 26) | θ value | 0.00175202 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG2_MOUSE
|
||||||
| NC score | 0.433186 (rank : 27) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_HUMAN
|
||||||
| NC score | 0.430817 (rank : 28) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
UNG2_HUMAN
|
||||||
| NC score | 0.353582 (rank : 29) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.103151 (rank : 30) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.084229 (rank : 31) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNL2_MOUSE
|
||||||
| NC score | 0.078447 (rank : 32) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.072138 (rank : 33) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL1_MOUSE
|
||||||
| NC score | 0.072003 (rank : 34) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL2_HUMAN
|
||||||
| NC score | 0.068863 (rank : 35) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
IER2_HUMAN
|
||||||
| NC score | 0.029588 (rank : 36) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTL4, Q03827, Q2TAZ2 | Gene names | IER2, ETR101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 2 protein (Protein ETR101). | |||||
|
NHRF2_HUMAN
|
||||||
| NC score | 0.022749 (rank : 37) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.022171 (rank : 38) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
GGPPS_MOUSE
|
||||||
| NC score | 0.016657 (rank : 39) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTN0 | Gene names | Ggps1 | |||
|
Domain Architecture |
|
|||||
| Description | Geranylgeranyl pyrophosphate synthetase (GGPP synthetase) (GGPPSase) (Geranylgeranyl diphosphate synthase) [Includes: Dimethylallyltranstransferase (EC 2.5.1.1); Geranyltranstransferase (EC 2.5.1.10); Farnesyltranstransferase (EC 2.5.1.29)]. | |||||
|
BUB1B_MOUSE
|
||||||
| NC score | 0.012350 (rank : 40) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
DOCK1_HUMAN
|
||||||
| NC score | 0.010229 (rank : 41) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14185 | Gene names | DOCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180). | |||||
|
ZWINT_MOUSE
|
||||||
| NC score | 0.006218 (rank : 42) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.005842 (rank : 43) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
SAFB2_MOUSE
|
||||||
| NC score | 0.005299 (rank : 44) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
LMNB2_HUMAN
|
||||||
| NC score | 0.004623 (rank : 45) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
TLR7_HUMAN
|
||||||
| NC score | 0.003381 (rank : 46) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYK1, Q9NR98 | Gene names | TLR7 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 7 precursor. | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.003070 (rank : 47) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.000812 (rank : 48) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
M3K4_MOUSE
|
||||||
| NC score | -0.000144 (rank : 49) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
ZBTB5_MOUSE
|
||||||
| NC score | -0.002615 (rank : 50) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 801 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQG0, Q3TVR7, Q6GV09, Q8BIB5 | Gene names | Zbtb5, Kiaa0354 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 5 (Transcription factor ZNF-POZ). | |||||