Please be patient as the page loads
|
STA5B_MOUSE
|
||||||
| SwissProt Accessions | P42232, Q541Q5, Q60804, Q8K3Q1, Q9JKM1 | Gene names | Stat5b | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STA5A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993426 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P42229 | Gene names | STAT5A, STAT5 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A. | |||||
|
STA5A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995968 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
STA5B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.997104 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P51692, Q8WWS8 | Gene names | STAT5B | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | P42232, Q541Q5, Q60804, Q8K3Q1, Q9JKM1 | Gene names | Stat5b | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STAT6_MOUSE
|
||||||
| θ value | 3.64273e-126 (rank : 5) | NC score | 0.944470 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
STAT6_HUMAN
|
||||||
| θ value | 1.80787e-125 (rank : 6) | NC score | 0.938051 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
STAT4_HUMAN
|
||||||
| θ value | 6.95003e-85 (rank : 7) | NC score | 0.896637 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT4_MOUSE
|
||||||
| θ value | 1.18549e-84 (rank : 8) | NC score | 0.900733 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT3_HUMAN
|
||||||
| θ value | 2.02215e-84 (rank : 9) | NC score | 0.890912 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P40763, O14916, Q9BW54 | Gene names | STAT3, APRF | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT3_MOUSE
|
||||||
| θ value | 2.02215e-84 (rank : 10) | NC score | 0.890910 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P42227 | Gene names | Stat3, Aprf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT1_HUMAN
|
||||||
| θ value | 5.33382e-77 (rank : 11) | NC score | 0.886793 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
STAT1_MOUSE
|
||||||
| θ value | 4.99229e-75 (rank : 12) | NC score | 0.885436 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P42225 | Gene names | Stat1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1. | |||||
|
STAT2_HUMAN
|
||||||
| θ value | 4.39379e-55 (rank : 13) | NC score | 0.873324 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 6.7944e-48 (rank : 14) | NC score | 0.818896 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 15) | NC score | 0.095980 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 16) | NC score | 0.081550 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 17) | NC score | 0.087997 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 18) | NC score | 0.075445 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 19) | NC score | 0.085826 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 20) | NC score | 0.089263 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.079374 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.075748 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.039312 (rank : 160) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.076741 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.072163 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.046229 (rank : 153) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
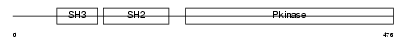
SH22A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.116736 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.071667 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.056923 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.040255 (rank : 158) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.077047 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.071617 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.088320 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.056023 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.079382 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.068240 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
HSH2D_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.071407 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JZ2, Q6ZNG7 | Gene names | HSH2D, ALX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
P55G_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.073224 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P55G_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.078804 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.001968 (rank : 188) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.071096 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.074290 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.065594 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.074021 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
FOXP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.036832 (rank : 164) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15409, Q6ZND1, Q75MJ3, Q8IZE0, Q8N0W2, Q8N6B7, Q8N6B8, Q8NFQ1, Q8NFQ2, Q8NFQ3, Q8NFQ4, Q8TD74 | Gene names | FOXP2, CAGH44, TNRC10 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P2 (CAG repeat protein 44) (Trinucleotide repeat- containing gene 10 protein). | |||||
|
FOXP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.037654 (rank : 163) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P58463, Q6PD37, Q8C4F0, Q8R441 | Gene names | Foxp2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P2. | |||||
|
HAPIP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.048531 (rank : 145) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.022867 (rank : 174) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.029496 (rank : 168) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.024351 (rank : 172) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.081775 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.054772 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.062719 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.048200 (rank : 147) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.049082 (rank : 143) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.068257 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CHIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.023127 (rank : 173) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.067788 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
HSH2D_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.084913 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.063135 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
XE7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.054701 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.065020 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
DNMBP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.017247 (rank : 181) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
K22E_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.020481 (rank : 178) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.016016 (rank : 182) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.052990 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.018118 (rank : 179) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.057882 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
BAIP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.017985 (rank : 180) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UQB8, O43858, Q53HB1, Q86WC1, Q8N5C0, Q96CR7, Q9UBR3, Q9UQ43 | Gene names | BAIAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2 (BAI1- associated protein 2) (BAI-associated protein 2) (Protein BAP2) (Insulin receptor substrate p53) (IRSp53) (Insulin receptor substrate protein of 53 kDa) (Insulin receptor substrate p53/p58) (IRSp53/58) (IRS-58) (Fas ligand-associated factor 3) (FLAF3). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.056584 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.052548 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CHIN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.020533 (rank : 177) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.065121 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.063057 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.058428 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.039803 (rank : 159) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.046857 (rank : 149) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.014954 (rank : 183) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.053031 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.039128 (rank : 161) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
P85A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.057320 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.055557 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.053571 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.041668 (rank : 156) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
TUFT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.057418 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.067049 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.046592 (rank : 151) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.063335 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
EVC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.026213 (rank : 169) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P57680 | Gene names | Evc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein homolog. | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.060867 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
K1683_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.040950 (rank : 157) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.036727 (rank : 165) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MED12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.044625 (rank : 154) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
P85B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.069419 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PPT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.021633 (rank : 175) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMR5, O14799, Q0P6K0, Q5JP13, Q5JP14, Q5JQF0, Q5SSX4, Q5SSX5, Q5SSX6, Q5STJ4, Q5STJ5, Q5STJ6, Q6FI80, Q99945 | Gene names | PPT2 | |||
|
Domain Architecture |
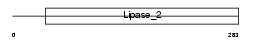
|
|||||
| Description | Lysosomal thioesterase PPT2 precursor (EC 3.1.2.-) (PPT-2) (S- thioesterase G14). | |||||
|
RABE2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.060567 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.051373 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CASP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.050295 (rank : 138) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CR034_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.054406 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.046384 (rank : 152) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.055870 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.043719 (rank : 155) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.010521 (rank : 185) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.047074 (rank : 148) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.066802 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
P85B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.070009 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
SOCS4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.036679 (rank : 166) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXH5 | Gene names | SOCS4, SOCS7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
SOCS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.039022 (rank : 162) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZA6 | Gene names | Socs4, Socs7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.003403 (rank : 187) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.058515 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
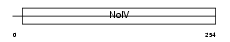
ANR26_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.048491 (rank : 146) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
APOA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.046631 (rank : 150) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |
|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.020752 (rank : 176) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.034247 (rank : 167) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
SRMS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.001748 (rank : 189) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||
|
APOE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.048739 (rank : 144) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P08226 | Gene names | Apoe | |||
|
Domain Architecture |
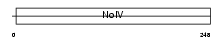
|
|||||
| Description | Apolipoprotein E precursor (Apo-E). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.052843 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.066113 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.066276 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
PAR14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.008090 (rank : 186) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
SH22A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.063831 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
SOCS5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.025754 (rank : 171) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75159, Q8IYZ4 | Gene names | SOCS5, CIS6, CISH5, CISH6, KIAA0671 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5) (Cytokine-inducible SH2 protein 6) (CIS-6). | |||||
|
SOCS5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.026034 (rank : 170) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54928 | Gene names | Socs5, Cish5 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5). | |||||
|
UCHL5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.010599 (rank : 184) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5K5, Q5LJA6, Q5LJA7, Q8TBS4, Q96BJ9, Q9H1W5, Q9P0I3, Q9UQN2 | Gene names | UCHL5, UCH37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.050536 (rank : 131) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
AZI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052822 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.054926 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
C102B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.050053 (rank : 141) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
CCD11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.052454 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CCD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.050342 (rank : 136) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CCD19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.059781 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.055407 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.074461 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CCD27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.060993 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CCD39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.050462 (rank : 133) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.056253 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.052778 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.055675 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.050859 (rank : 129) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CE152_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.050146 (rank : 140) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CE290_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.055761 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CEP55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.050019 (rank : 142) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.051337 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CING_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.051367 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CING_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.055839 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CJ118_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.051753 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.050943 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CKAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.050747 (rank : 130) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.052243 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.056374 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.057534 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DESP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.051021 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.055867 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.053965 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.052272 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
GCC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.056010 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.052093 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.060591 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.062538 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.050902 (rank : 128) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.052023 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.051956 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.053436 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.053452 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.050491 (rank : 132) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
INVO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.055883 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
KTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.053506 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.050442 (rank : 134) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
NAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.055770 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.056203 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
NEMO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.055551 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
NIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.058665 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
NIN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.055435 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PCNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.061990 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PEPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.050323 (rank : 137) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PININ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.050417 (rank : 135) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.054570 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.055081 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.057208 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.054537 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.056353 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
REST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.052382 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
REST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.061006 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.057034 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.056662 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.051938 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.050968 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.050231 (rank : 139) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.065446 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
STA5B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | P42232, Q541Q5, Q60804, Q8K3Q1, Q9JKM1 | Gene names | Stat5b | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5B_HUMAN
|
||||||
| NC score | 0.997104 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P51692, Q8WWS8 | Gene names | STAT5B | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5B. | |||||
|
STA5A_MOUSE
|
||||||
| NC score | 0.995968 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P42230 | Gene names | Stat5a, Mgf, Mpf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A (Mammary gland factor). | |||||
|
STA5A_HUMAN
|
||||||
| NC score | 0.993426 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P42229 | Gene names | STAT5A, STAT5 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 5A. | |||||
|
STAT6_MOUSE
|
||||||
| NC score | 0.944470 (rank : 5) | θ value | 3.64273e-126 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
STAT6_HUMAN
|
||||||
| NC score | 0.938051 (rank : 6) | θ value | 1.80787e-125 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
STAT4_MOUSE
|
||||||
| NC score | 0.900733 (rank : 7) | θ value | 1.18549e-84 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P42228 | Gene names | Stat4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT4_HUMAN
|
||||||
| NC score | 0.896637 (rank : 8) | θ value | 6.95003e-85 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14765, Q96NZ6 | Gene names | STAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 4. | |||||
|
STAT3_HUMAN
|
||||||
| NC score | 0.890912 (rank : 9) | θ value | 2.02215e-84 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P40763, O14916, Q9BW54 | Gene names | STAT3, APRF | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT3_MOUSE
|
||||||
| NC score | 0.890910 (rank : 10) | θ value | 2.02215e-84 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P42227 | Gene names | Stat3, Aprf | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 3 (Acute-phase response factor). | |||||
|
STAT1_HUMAN
|
||||||
| NC score | 0.886793 (rank : 11) | θ value | 5.33382e-77 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
STAT1_MOUSE
|
||||||
| NC score | 0.885436 (rank : 12) | θ value | 4.99229e-75 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P42225 | Gene names | Stat1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1. | |||||
|
STAT2_HUMAN
|
||||||
| NC score | 0.873324 (rank : 13) | θ value | 4.39379e-55 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.818896 (rank : 14) | θ value | 6.7944e-48 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
SH22A_HUMAN
|
||||||
| NC score | 0.116736 (rank : 15) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.095980 (rank : 16) | θ value | 5.44631e-05 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.089263 (rank : 17) | θ value | 0.0148317 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.088320 (rank : 18) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.087997 (rank : 19) | θ value | 0.00390308 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.085826 (rank : 20) | θ value | 0.00869519 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
HSH2D_MOUSE
|
||||||
| NC score | 0.084913 (rank : 21) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.081775 (rank : 22) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.081550 (rank : 23) | θ value | 0.00298849 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.079382 (rank : 24) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.079374 (rank : 25) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
P55G_MOUSE
|
||||||
| NC score | 0.078804 (rank : 26) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.077047 (rank : 27) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.076741 (rank : 28) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.075748 (rank : 29) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.075445 (rank : 30) | θ value | 0.00869519 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD27_HUMAN
|
||||||
| NC score | 0.074461 (rank : 31) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.074290 (rank : 32) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.074021 (rank : 33) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
P55G_HUMAN
|
||||||
| NC score | 0.073224 (rank : 34) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.072163 (rank : 35) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.071667 (rank : 36) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.071617 (rank : 37) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
HSH2D_HUMAN
|
||||||
| NC score | 0.071407 (rank : 38) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JZ2, Q6ZNG7 | Gene names | HSH2D, ALX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.071096 (rank : 39) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.070009 (rank : 40) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.069419 (rank : 41) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.068257 (rank : 42) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.068240 (rank : 43) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.067788 (rank : 44) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.067049 (rank : 45) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.066802 (rank : 46) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.066276 (rank : 47) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.066113 (rank : 48) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.065594 (rank : 49) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.065446 (rank : 50) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.065121 (rank : 51) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.065020 (rank : 52) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
SH22A_MOUSE
|
||||||
| NC score | 0.063831 (rank : 53) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.063335 (rank : 54) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.063135 (rank : 55) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.063057 (rank : 56) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.062719 (rank : 57) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.062538 (rank : 58) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.061990 (rank : 59) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.061006 (rank : 60) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
CCD27_MOUSE
|
||||||
| NC score | 0.060993 (rank : 61) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.060867 (rank : 62) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.060591 (rank : 63) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
RABE2_MOUSE
|
||||||
| NC score | 0.060567 (rank : 64) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
CCD19_HUMAN
|
||||||
| NC score | 0.059781 (rank : 65) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.058665 (rank : 66) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.058515 (rank : 67) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.058428 (rank : 68) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.057882 (rank : 69) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.057534 (rank : 70) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
TUFT1_HUMAN
|
||||||
| NC score | 0.057418 (rank : 71) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.057320 (rank : 72) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.057208 (rank : 73) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.057034 (rank : 74) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.056923 (rank : 75) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.056662 (rank : 76) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.056584 (rank : 77) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.056374 (rank : 78) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.056353 (rank : 79) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.056253 (rank : 80) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.056203 (rank : 81) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.056023 (rank : 82) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.056010 (rank : 83) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.055883 (rank : 84) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.055870 (rank : 85) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.055867 (rank : 86) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.055839 (rank : 87) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.055770 (rank : 88) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.055761 (rank : 89) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.055675 (rank : 90) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.055557 (rank : 91) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
NEMO_MOUSE
|
||||||
| NC score | 0.055551 (rank : 92) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.055435 (rank : 93) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.055407 (rank : 94) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.055081 (rank : 95) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.054926 (rank : 96) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.054772 (rank : 97) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.054701 (rank : 98) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.054570 (rank : 99) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.054537 (rank : 100) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
CR034_HUMAN
|
||||||
| NC score | 0.054406 (rank : 101) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.053965 (rank : 102) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.053571 (rank : 103) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.053506 (rank : 104) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.053452 (rank : 105) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.053436 (rank : 106) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.053031 (rank : 107) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.052990 (rank : 108) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.052843 (rank : 109) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.052822 (rank : 110) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.052778 (rank : 111) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.052548 (rank : 112) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CCD11_MOUSE
|
||||||
| NC score | 0.052454 (rank : 113) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.052382 (rank : 114) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.052272 (rank : 115) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.052243 (rank : 116) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.052093 (rank : 117) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.052023 (rank : 118) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.051956 (rank : 119) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.051938 (rank : 120) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.051753 (rank : 121) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.051373 (rank : 122) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.051367 (rank : 123) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.051337 (rank : 124) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.051021 (rank : 125) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.050968 (rank : 126) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.050943 (rank : 127) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.050902 (rank : 128) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.050859 (rank : 129) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CKAP4_MOUSE
|
||||||
| NC score | 0.050747 (rank : 130) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.050536 (rank : 131) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.050491 (rank : 132) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.050462 (rank : 133) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.050442 (rank : 134) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.050417 (rank : 135) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.050342 (rank : 136) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.050323 (rank : 137) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.050295 (rank : 138) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.050231 (rank : 139) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.050146 (rank : 140) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
C102B_HUMAN
|
||||||
| NC score | 0.050053 (rank : 141) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
CEP55_HUMAN
|
||||||
| NC score | 0.050019 (rank : 142) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.049082 (rank : 143) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
APOE_MOUSE
|
||||||
| NC score | 0.048739 (rank : 144) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P08226 | Gene names | Apoe | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein E precursor (Apo-E). | |||||
|
HAPIP_HUMAN
|
||||||
| NC score | 0.048531 (rank : 145) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.048491 (rank : 146) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.048200 (rank : 147) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.047074 (rank : 148) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.046857 (rank : 149) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
APOA4_MOUSE
|
||||||
| NC score | 0.046631 (rank : 150) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.046592 (rank : 151) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.046384 (rank : 152) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.046229 (rank : 153) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MED12_HUMAN
|
||||||
| NC score | 0.044625 (rank : 154) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.043719 (rank : 155) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.041668 (rank : 156) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.040950 (rank : 157) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.040255 (rank : 158) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.039803 (rank : 159) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.039312 (rank : 160) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.039128 (rank : 161) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
SOCS4_MOUSE
|
||||||
| NC score | 0.039022 (rank : 162) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZA6 | Gene names | Socs4, Socs7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
FOXP2_MOUSE
|
||||||
| NC score | 0.037654 (rank : 163) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P58463, Q6PD37, Q8C4F0, Q8R441 | Gene names | Foxp2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P2. | |||||
|
FOXP2_HUMAN
|
||||||
| NC score | 0.036832 (rank : 164) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15409, Q6ZND1, Q75MJ3, Q8IZE0, Q8N0W2, Q8N6B7, Q8N6B8, Q8NFQ1, Q8NFQ2, Q8NFQ3, Q8NFQ4, Q8TD74 | Gene names | FOXP2, CAGH44, TNRC10 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P2 (CAG repeat protein 44) (Trinucleotide repeat- containing gene 10 protein). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.036727 (rank : 165) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
SOCS4_HUMAN
|
||||||
| NC score | 0.036679 (rank : 166) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXH5 | Gene names | SOCS4, SOCS7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.034247 (rank : 167) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.029496 (rank : 168) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
EVC_MOUSE
|
||||||
| NC score | 0.026213 (rank : 169) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P57680 | Gene names | Evc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein homolog. | |||||
|
SOCS5_MOUSE
|
||||||
| NC score | 0.026034 (rank : 170) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54928 | Gene names | Socs5, Cish5 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5). | |||||
|
SOCS5_HUMAN
|
||||||
| NC score | 0.025754 (rank : 171) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75159, Q8IYZ4 | Gene names | SOCS5, CIS6, CISH5, CISH6, KIAA0671 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 5 (SOCS-5) (Cytokine-inducible SH2- containing protein 5) (Cytokine-inducible SH2 protein 6) (CIS-6). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.024351 (rank : 172) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
CHIN_HUMAN
|
||||||
| NC score | 0.023127 (rank : 173) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.022867 (rank : 174) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
PPT2_HUMAN
|
||||||
| NC score | 0.021633 (rank : 175) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMR5, O14799, Q0P6K0, Q5JP13, Q5JP14, Q5JQF0, Q5SSX4, Q5SSX5, Q5SSX6, Q5STJ4, Q5STJ5, Q5STJ6, Q6FI80, Q99945 | Gene names | PPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal thioesterase PPT2 precursor (EC 3.1.2.-) (PPT-2) (S- thioesterase G14). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.020752 (rank : 176) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
CHIN_MOUSE
|
||||||
| NC score | 0.020533 (rank : 177) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
K22E_HUMAN
|
||||||
| NC score | 0.020481 (rank : 178) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.018118 (rank : 179) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
BAIP2_HUMAN
|
||||||
| NC score | 0.017985 (rank : 180) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UQB8, O43858, Q53HB1, Q86WC1, Q8N5C0, Q96CR7, Q9UBR3, Q9UQ43 | Gene names | BAIAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2 (BAI1- associated protein 2) (BAI-associated protein 2) (Protein BAP2) (Insulin receptor substrate p53) (IRSp53) (Insulin receptor substrate protein of 53 kDa) (Insulin receptor substrate p53/p58) (IRSp53/58) (IRS-58) (Fas ligand-associated factor 3) (FLAF3). | |||||
|
DNMBP_HUMAN
|
||||||
| NC score | 0.017247 (rank : 181) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.016016 (rank : 182) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.014954 (rank : 183) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
UCHL5_HUMAN
|
||||||
| NC score | 0.010599 (rank : 184) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5K5, Q5LJA6, Q5LJA7, Q8TBS4, Q96BJ9, Q9H1W5, Q9P0I3, Q9UQN2 | Gene names | UCHL5, UCH37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.010521 (rank : 185) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PAR14_MOUSE
|
||||||
| NC score | 0.008090 (rank : 186) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.003403 (rank : 187) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.001968 (rank : 188) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SRMS_HUMAN
|
||||||
| NC score | 0.001748 (rank : 189) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||