Please be patient as the page loads
|
PARC_HUMAN
|
||||||
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CUL7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.794754 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
PARC_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.986191 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
ARI2_HUMAN
|
||||||
| θ value | 2.67181e-44 (rank : 4) | NC score | 0.704588 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95376, Q9HBZ6, Q9UEM9 | Gene names | ARIH2, ARI2, TRIAD1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-2 homolog (ARI-2) (Triad1 protein). | |||||
|
ARI2_MOUSE
|
||||||
| θ value | 4.55743e-44 (rank : 5) | NC score | 0.704003 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z1K6 | Gene names | Arih2, Ari2, Triad1, Uip48 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-2 homolog (ARI-2) (Triad1 protein) (UbcM4-interacting protein 48). | |||||
|
ARI1_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 6) | NC score | 0.688363 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y4X5, O76026, Q9H3T6, Q9UEN0, Q9UP39 | Gene names | ARIH1, ARI, UBCH7BP | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein) (HHARI) (H7-AP2) (MOP-6). | |||||
|
ARI1_MOUSE
|
||||||
| θ value | 2.26708e-35 (rank : 7) | NC score | 0.688230 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z1K5, Q6PF92 | Gene names | Arih1, Ari, Ubch7bp, Uip77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein 77). | |||||
|
HECD3_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 8) | NC score | 0.356604 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 9) | NC score | 0.260986 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HECD3_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 10) | NC score | 0.356785 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
RNF14_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 11) | NC score | 0.592242 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBS8, O94793, Q6IBV0 | Gene names | RNF14, ARA54 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 14 (Androgen receptor-associated protein 54) (Triad2 protein) (HFB30). | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 12) | NC score | 0.254636 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
RNF14_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 13) | NC score | 0.593726 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JI90, Q9D0L2, Q9D6N2, Q9D6Z8, Q9JI89 | Gene names | Rnf14, Ara54, Triad2 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 14 (Androgen receptor-associated protein 54) (Triad2 protein). | |||||
|
IBRD3_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 14) | NC score | 0.567469 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6ZMZ0, Q6P6A4 | Gene names | IBRDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IBR domain-containing protein 3. | |||||
|
RNF19_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 15) | NC score | 0.535592 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NV58, Q9H5H9, Q9H8M8, Q9UFG0, Q9UFX6, Q9Y4Y1 | Gene names | RNF19 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (Dorfin) (Double ring-finger protein) (p38 protein). | |||||
|
RNF19_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 16) | NC score | 0.537188 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50636, Q9QUJ5 | Gene names | Rnf19, Geg-154, Xybp | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (XY body protein) (XYbp) (Gametogenesis expressed protein GEG-154) (UBCM4-interacting protein 117) (UIP117). | |||||
|
UB7I4_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 17) | NC score | 0.572828 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q925F3, Q6A0B4 | Gene names | Rnf144, Kiaa0161, Ubce7ip4, Uip4 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
UB7I4_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 18) | NC score | 0.569648 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P50876 | Gene names | RNF144, KIAA0161, UBCE7IP4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
IBRD2_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 19) | NC score | 0.551799 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BKD6, Q8BG97 | Gene names | Ibrdc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase IBRDC2 (EC 6.3.2.-) (IBR domain-containing protein 2). | |||||
|
PRKN2_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 20) | NC score | 0.499873 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
PRKN2_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 21) | NC score | 0.501651 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVS6, Q9ES22, Q9ES23 | Gene names | Park2, Prkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN). | |||||
|
IBRD2_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 22) | NC score | 0.539059 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z419, Q6P4Q0, Q8N3R7, Q9BX38 | Gene names | IBRDC2, P53RFP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase IBRDC2 (EC 6.3.2.-) (IBR domain-containing protein 2) (p53-inducible RING finger protein). | |||||
|
RNF31_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 23) | NC score | 0.420591 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
RNF31_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 24) | NC score | 0.423928 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
UB7I3_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 25) | NC score | 0.441749 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
UB7I3_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 26) | NC score | 0.439187 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BYM8, O95623, Q96BS3, Q9BYM9 | Gene names | RBCK1, C20orf18, RNF54, UBCE7IP3, XAP4 | |||
|
Domain Architecture |
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (Hepatitis B virus X- associated protein 4) (HBV-associated factor 4) (RING finger protein 54). | |||||
|
IBRD1_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 27) | NC score | 0.533604 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TC41, Q9BX48 | Gene names | IBRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IBR domain-containing protein 1. | |||||
|
CUL4B_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 28) | NC score | 0.120236 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CUL4A_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 29) | NC score | 0.118158 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
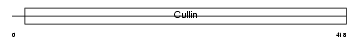
CUL4A_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 30) | NC score | 0.118721 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |
|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
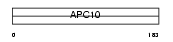
APC10_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 31) | NC score | 0.274878 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UM13, Q2V500, Q9UG51, Q9Y5R0 | Gene names | ANAPC10, APC10 | |||
|
Domain Architecture |
|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
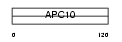
APC10_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 32) | NC score | 0.272998 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2H6, Q9CW24 | Gene names | Anapc10, Apc10 | |||
|
Domain Architecture |
|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
CUL3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.084673 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 34) | NC score | 0.084827 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
PDE9A_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 35) | NC score | 0.040607 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
UB7I1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 36) | NC score | 0.309899 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
NOL8_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 37) | NC score | 0.041338 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
PDE9A_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 38) | NC score | 0.037135 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 39) | NC score | 0.307020 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
INHA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.033060 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05111 | Gene names | INHA | |||
|
Domain Architecture |

|
|||||
| Description | Inhibin alpha chain precursor. | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.062314 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.059514 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
FBN3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 43) | NC score | 0.017827 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.013627 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
GOLI_HUMAN
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.051723 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86XS8, Q641P9, Q6P177, Q7L2U2, Q9P0J9 | Gene names | RNF130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130). | |||||
|
GOLI_MOUSE
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.043094 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VEM1, Q80VL7, Q9QZQ6 | Gene names | Rnf130, G1rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130) (G1-related zinc finger protein). | |||||
|
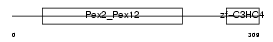
PEX10_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.071111 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60683, Q5T095, Q9BW90 | Gene names | PEX10, RNF69 | |||
|
Domain Architecture |
|
|||||
| Description | Peroxisome assembly protein 10 (Peroxin-10) (Peroxisome biogenesis factor 10) (RING finger protein 69). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.006540 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
TRIM9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.042449 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C026, Q92557, Q96D24, Q96NI4, Q9C025, Q9C027 | Gene names | TRIM9, KIAA0282, RNF91 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9 (RING finger protein 91). | |||||
|
CD177_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.054984 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6Q3, Q711Q2, Q8NCV9, Q96QH1, Q9HDA5 | Gene names | CD177, NB1, PRV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD177 antigen precursor (Polycythemia rubra vera protein 1) (PRV-1) (NB1 glycoprotein) (NB1 GP) (Human neutrophil alloantigen 2a) (HNA- 2a). | |||||
|
OST3B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.012538 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZS6 | Gene names | Hs3st3b1, 3ost3b1, Hs3st3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (m3-OST-3B). | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.070819 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
TOPRS_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.072155 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.022971 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.013402 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
CLUA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.032627 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96AJ1, O75138, Q65ZA3, Q9H8R4, Q9H8T1 | Gene names | CLUAP1, KIAA0643 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
CLUA1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.032302 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
STX4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.016402 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12846, Q15525 | Gene names | STX4, STX4A | |||
|
Domain Architecture |
|
|||||
| Description | Syntaxin-4 (NY-REN-31 antigen). | |||||
|
ADA18_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.011975 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R157, Q60621 | Gene names | Adam18, Adam27, Dtgn3, Tmdc3 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 18 precursor (A disintegrin and metalloproteinase domain 18) (Transmembrane metalloproteinase-like, disintegrin-like, and cysteine- rich protein III) (tMDC III) (ADAM 27). | |||||
|
B3A4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.010983 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96Q91, Q96RM5, Q9BXF2, Q9BXN3 | Gene names | SLC4A9, AE4, SBC5 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 4 (Anion exchanger 4) (Solute carrier family 4 member 9) (Sodium bicarbonate cotransporter 5). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.017502 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
JIP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.013182 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
PCSK5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.009853 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
IPO9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.027979 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96P70, Q8N1Y1, Q8N3I2, Q8NCG9, Q96SU6, Q9NW01, Q9P0A8, Q9ULM8 | Gene names | IPO9, IMP9, KIAA1192, RANBP9 | |||
|
Domain Architecture |
|
|||||
| Description | Importin-9 (Imp9) (Ran-binding protein 9) (RanbP9). | |||||
|
IPO9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.027916 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE6, Q924M3, Q924M4 | Gene names | Ipo9, Imp9, Ranbp9 | |||
|
Domain Architecture |
|
|||||
| Description | Importin-9 (Importin 9a) (Imp9a) (Importin 9b) (Imp9b) (Ran-binding protein 9) (RanBP9). | |||||
|
MICA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.009456 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.002397 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
RNF6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.025148 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y252, Q9UF41 | Gene names | RNF6 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 6 (RING-H2 protein). | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.014529 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ARHGC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.008537 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ARMC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.021050 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.016201 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
KR104_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.013940 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.007363 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
RN185_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.046787 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96GF1, Q8N900 | Gene names | RNF185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
RN185_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.045896 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91YT2, Q6ZWS3 | Gene names | Rnf185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
STX4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.014659 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70452 | Gene names | Stx4, Stx4a | |||
|
Domain Architecture |
|
|||||
| Description | Syntaxin-4. | |||||
|
TF65_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.013195 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.011372 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.009120 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
CF049_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.015092 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2TBC4, Q5T3D4, Q9NSV1 | Gene names | C6orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf49. | |||||
|
KR105_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.015150 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.007552 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PPR3D_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.026264 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95685, Q6DK02 | Gene names | PPP1R3D, PPP1R6 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 3D (Protein phosphatase 1, regulatory subunit 6) (Protein phosphatase 1-binding subunit R6). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.013683 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
TRIM9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.036595 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C7M3, Q6ZQE5, Q80WT6, Q8C7Z4, Q8CC32, Q8CEG2, Q99PQ3 | Gene names | Trim9, Kiaa0282 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9. | |||||
|
AAKG3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.011004 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGM7, Q80WK8, Q8CJ41 | Gene names | Prkag3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-3 (AMPK gamma-3 chain) (AMPK gamma3). | |||||
|
ATS13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.004566 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CF182_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.009647 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
KRA58_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.011358 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.033149 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
PLCB3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.008062 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
PLXA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.006802 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.007697 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RNF5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.049828 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35445 | Gene names | Rnf5, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (RING finger protein 5). | |||||
|
RTN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.013913 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70622, O70620 | Gene names | Rtn2, Nspl1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
THUM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.017396 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99J36, Q3U6Y3 | Gene names | Thumpd1 | |||
|
Domain Architecture |

|
|||||
| Description | THUMP domain-containing protein 1. | |||||
|
ABLM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.013120 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.007839 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.010390 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
EGFL7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.010156 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UHF1, Q5M7Y5, Q5VUD5, Q96EG0 | Gene names | EGFL7, MEGF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 7 precursor (Multiple EGF-like domain protein 7) (Multiple epidermal growth factor-like domain protein 7) (Vascular endothelial statin) (VE-statin) (NOTCH4-like protein) (ZNEU1). | |||||
|
ELL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.013053 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80VR2 | Gene names | Ell3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.012267 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
FAS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.009548 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.012782 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.005038 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.032506 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.034100 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
NBN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.012802 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
PYGO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.013007 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0P5 | Gene names | Pygo1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.023253 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.021408 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TAOK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | -0.002035 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1494 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H2K8, Q658N1, Q8IUM4, Q9HC79, Q9NZM9, Q9UHG7 | Gene names | TAOK3, DPK, JIK, KDS, MAP3K18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3) (Jun kinase-inhibitory kinase) (JNK/SAPK- inhibitory kinase) (Dendritic cell-derived protein kinase) (Cutaneous T-cell lymphoma tumor antigen HD-CL-09) (CTCL tumor antigen HD-CL-09) (Kinase from chicken homolog A) (hKFC-A). | |||||
|
TAOK3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | -0.001888 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.039774 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.036587 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
NC6IP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.011169 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RS0, Q5GH23, Q8TDG9, Q96QU3, Q9H5V3 | Gene names | NCOA6IP, PIMT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT) (CLL-associated antigen KW-2) (Hepatocellular carcinoma-associated antigen 137) (HCA137). | |||||
|
PA2G3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.016999 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ20, O95768 | Gene names | PLA2G3 | |||
|
Domain Architecture |
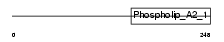
|
|||||
| Description | Group 3 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group III secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GIII) (GIII sPLA2). | |||||
|
PACN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.004217 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.002710 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
RN141_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.025274 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WVD5, Q9NZB4 | Gene names | RNF141, ZNF230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 141 (Zinc finger protein 230). | |||||
|
RNF5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.046285 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99942, Q9UMQ2 | Gene names | RNF5, G16, NG2, RMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (HsRma1) (RING finger protein 5) (Protein G16). | |||||
|
SON_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.005418 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TMPS6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.000107 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
ASPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.011841 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.005386 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.000347 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.002471 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CT077_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.007703 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQG5 | Gene names | C20orf77 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf77. | |||||
|
FBN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.008497 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.000102 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
IGF1R_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | -0.002797 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
KRA24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.012794 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.008250 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.008016 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.011282 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
NFKB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.003824 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19838, Q68D84, Q86V43, Q8N4X7, Q9NZC0 | Gene names | NFKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) [Contains: Nuclear factor NF-kappa-B p50 subunit]. | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.006819 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.010157 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.005487 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
RN122_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.026557 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H9V4, Q52LK3 | Gene names | RNF122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
RN122_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.024303 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BP31, Q80VA7, Q8BGD3 | Gene names | Rnf122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
RN141_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.023941 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99MB7, Q9D040, Q9D5C4, Q9D9L0 | Gene names | Rnf141, Znf230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 141 (Zinc finger protein 230). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.003766 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
SYT14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.004446 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.002980 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | -0.003790 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
ZN425_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | -0.003975 (rank : 163) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6IV72 | Gene names | ZNF425 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 425. | |||||
|
CUL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.077021 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.077015 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.050572 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13617, O00200, Q9UNF9 | Gene names | CUL2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050457 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D4H8, Q3TUR8 | Gene names | Cul2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.052536 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.052612 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.050269 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HERC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.051437 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HERC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.052672 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.050942 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.050898 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.056386 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.051460 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.053263 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.052934 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
PARC_MOUSE
|
||||||
| NC score | 0.986191 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.794754 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
ARI2_HUMAN
|
||||||
| NC score | 0.704588 (rank : 4) | θ value | 2.67181e-44 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95376, Q9HBZ6, Q9UEM9 | Gene names | ARIH2, ARI2, TRIAD1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-2 homolog (ARI-2) (Triad1 protein). | |||||
|
ARI2_MOUSE
|
||||||
| NC score | 0.704003 (rank : 5) | θ value | 4.55743e-44 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z1K6 | Gene names | Arih2, Ari2, Triad1, Uip48 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-2 homolog (ARI-2) (Triad1 protein) (UbcM4-interacting protein 48). | |||||
|
ARI1_HUMAN
|
||||||
| NC score | 0.688363 (rank : 6) | θ value | 2.26708e-35 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y4X5, O76026, Q9H3T6, Q9UEN0, Q9UP39 | Gene names | ARIH1, ARI, UBCH7BP | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein) (HHARI) (H7-AP2) (MOP-6). | |||||
|
ARI1_MOUSE
|
||||||
| NC score | 0.688230 (rank : 7) | θ value | 2.26708e-35 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z1K5, Q6PF92 | Gene names | Arih1, Ari, Ubch7bp, Uip77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein 77). | |||||
|
RNF14_MOUSE
|
||||||
| NC score | 0.593726 (rank : 8) | θ value | 1.63225e-17 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JI90, Q9D0L2, Q9D6N2, Q9D6Z8, Q9JI89 | Gene names | Rnf14, Ara54, Triad2 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 14 (Androgen receptor-associated protein 54) (Triad2 protein). | |||||
|
RNF14_HUMAN
|
||||||
| NC score | 0.592242 (rank : 9) | θ value | 4.29542e-18 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBS8, O94793, Q6IBV0 | Gene names | RNF14, ARA54 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 14 (Androgen receptor-associated protein 54) (Triad2 protein) (HFB30). | |||||
|
UB7I4_MOUSE
|
||||||
| NC score | 0.572828 (rank : 10) | θ value | 2.20605e-14 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q925F3, Q6A0B4 | Gene names | Rnf144, Kiaa0161, Ubce7ip4, Uip4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
UB7I4_HUMAN
|
||||||
| NC score | 0.569648 (rank : 11) | θ value | 6.41864e-14 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P50876 | Gene names | RNF144, KIAA0161, UBCE7IP4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
IBRD3_HUMAN
|
||||||
| NC score | 0.567469 (rank : 12) | θ value | 4.02038e-16 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6ZMZ0, Q6P6A4 | Gene names | IBRDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IBR domain-containing protein 3. | |||||
|
IBRD2_MOUSE
|
||||||
| NC score | 0.551799 (rank : 13) | θ value | 1.33837e-11 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BKD6, Q8BG97 | Gene names | Ibrdc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase IBRDC2 (EC 6.3.2.-) (IBR domain-containing protein 2). | |||||
|
IBRD2_HUMAN
|
||||||
| NC score | 0.539059 (rank : 14) | θ value | 1.47974e-10 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z419, Q6P4Q0, Q8N3R7, Q9BX38 | Gene names | IBRDC2, P53RFP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase IBRDC2 (EC 6.3.2.-) (IBR domain-containing protein 2) (p53-inducible RING finger protein). | |||||
|
RNF19_MOUSE
|
||||||
| NC score | 0.537188 (rank : 15) | θ value | 8.95645e-16 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50636, Q9QUJ5 | Gene names | Rnf19, Geg-154, Xybp | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (XY body protein) (XYbp) (Gametogenesis expressed protein GEG-154) (UBCM4-interacting protein 117) (UIP117). | |||||
|
RNF19_HUMAN
|
||||||
| NC score | 0.535592 (rank : 16) | θ value | 8.95645e-16 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NV58, Q9H5H9, Q9H8M8, Q9UFG0, Q9UFX6, Q9Y4Y1 | Gene names | RNF19 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (Dorfin) (Double ring-finger protein) (p38 protein). | |||||
|
IBRD1_HUMAN
|
||||||
| NC score | 0.533604 (rank : 17) | θ value | 2.61198e-07 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TC41, Q9BX48 | Gene names | IBRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IBR domain-containing protein 1. | |||||
|
PRKN2_MOUSE
|
||||||
| NC score | 0.501651 (rank : 18) | θ value | 5.08577e-11 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVS6, Q9ES22, Q9ES23 | Gene names | Park2, Prkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN). | |||||
|
PRKN2_HUMAN
|
||||||
| NC score | 0.499873 (rank : 19) | θ value | 5.08577e-11 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
UB7I3_MOUSE
|
||||||
| NC score | 0.441749 (rank : 20) | θ value | 2.36244e-08 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
UB7I3_HUMAN
|
||||||
| NC score | 0.439187 (rank : 21) | θ value | 1.53129e-07 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BYM8, O95623, Q96BS3, Q9BYM9 | Gene names | RBCK1, C20orf18, RNF54, UBCE7IP3, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (Hepatitis B virus X- associated protein 4) (HBV-associated factor 4) (RING finger protein 54). | |||||
|
RNF31_HUMAN
|
||||||
| NC score | 0.423928 (rank : 22) | θ value | 1.80886e-08 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
RNF31_MOUSE
|
||||||
| NC score | 0.420591 (rank : 23) | θ value | 1.38499e-08 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.356785 (rank : 24) | θ value | 5.07402e-19 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.356604 (rank : 25) | θ value | 1.33526e-19 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
UB7I1_HUMAN
|
||||||
| NC score | 0.309899 (rank : 26) | θ value | 0.0252991 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.307020 (rank : 27) | θ value | 0.0961366 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
APC10_HUMAN
|
||||||
| NC score | 0.274878 (rank : 28) | θ value | 0.00228821 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UM13, Q2V500, Q9UG51, Q9Y5R0 | Gene names | ANAPC10, APC10 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
APC10_MOUSE
|
||||||
| NC score | 0.272998 (rank : 29) | θ value | 0.00228821 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2H6, Q9CW24 | Gene names | Anapc10, Apc10 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.260986 (rank : 30) | θ value | 1.74391e-19 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.254636 (rank : 31) | θ value | 5.60996e-18 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
CUL4B_HUMAN
|
||||||
| NC score | 0.120236 (rank : 32) | θ value | 4.1701e-05 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CUL4A_HUMAN
|
||||||
| NC score | 0.118721 (rank : 33) | θ value | 0.00175202 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4A_MOUSE
|
||||||
| NC score | 0.118158 (rank : 34) | θ value | 0.000602161 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL3_MOUSE
|
||||||
| NC score | 0.084827 (rank : 35) | θ value | 0.0252991 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_HUMAN
|
||||||
| NC score | 0.084673 (rank : 36) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL1_HUMAN
|
||||||
| NC score | 0.077021 (rank : 37) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| NC score | 0.077015 (rank : 38) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
TOPRS_MOUSE
|
||||||
| NC score | 0.072155 (rank : 39) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
PEX10_HUMAN
|
||||||
| NC score | 0.071111 (rank : 40) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60683, Q5T095, Q9BW90 | Gene names | PEX10, RNF69 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly protein 10 (Peroxin-10) (Peroxisome biogenesis factor 10) (RING finger protein 69). | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.070819 (rank : 41) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.062314 (rank : 42) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.059514 (rank : 43) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.056386 (rank : 44) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
CD177_HUMAN
|
||||||
| NC score | 0.054984 (rank : 45) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6Q3, Q711Q2, Q8NCV9, Q96QH1, Q9HDA5 | Gene names | CD177, NB1, PRV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD177 antigen precursor (Polycythemia rubra vera protein 1) (PRV-1) (NB1 glycoprotein) (NB1 GP) (Human neutrophil alloantigen 2a) (HNA- 2a). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.053263 (rank : 46) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.052934 (rank : 47) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.052672 (rank : 48) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.052612 (rank : 49) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.052536 (rank : 50) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
GOLI_HUMAN
|
||||||
| NC score | 0.051723 (rank : 51) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86XS8, Q641P9, Q6P177, Q7L2U2, Q9P0J9 | Gene names | RNF130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.051460 (rank : 52) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.051437 (rank : 53) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.050942 (rank : 54) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.050898 (rank : 55) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
CUL2_HUMAN
|
||||||
| NC score | 0.050572 (rank : 56) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13617, O00200, Q9UNF9 | Gene names | CUL2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL2_MOUSE
|
||||||
| NC score | 0.050457 (rank : 57) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D4H8, Q3TUR8 | Gene names | Cul2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.050269 (rank : 58) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
RNF5_MOUSE
|
||||||
| NC score | 0.049828 (rank : 59) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35445 | Gene names | Rnf5, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (RING finger protein 5). | |||||
|
RN185_HUMAN
|
||||||
| NC score | 0.046787 (rank : 60) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96GF1, Q8N900 | Gene names | RNF185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
RNF5_HUMAN
|
||||||
| NC score | 0.046285 (rank : 61) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99942, Q9UMQ2 | Gene names | RNF5, G16, NG2, RMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (HsRma1) (RING finger protein 5) (Protein G16). | |||||
|
RN185_MOUSE
|
||||||
| NC score | 0.045896 (rank : 62) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91YT2, Q6ZWS3 | Gene names | Rnf185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
GOLI_MOUSE
|
||||||
| NC score | 0.043094 (rank : 63) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VEM1, Q80VL7, Q9QZQ6 | Gene names | Rnf130, G1rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130) (G1-related zinc finger protein). | |||||
|
TRIM9_HUMAN
|
||||||
| NC score | 0.042449 (rank : 64) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C026, Q92557, Q96D24, Q96NI4, Q9C025, Q9C027 | Gene names | TRIM9, KIAA0282, RNF91 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9 (RING finger protein 91). | |||||
|
NOL8_MOUSE
|
||||||
| NC score | 0.041338 (rank : 65) | θ value | 0.0563607 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
PDE9A_MOUSE
|
||||||
| NC score | 0.040607 (rank : 66) | θ value | 0.0252991 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.039774 (rank : 67) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
PDE9A_HUMAN
|
||||||
| NC score | 0.037135 (rank : 68) | θ value | 0.0736092 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
TRIM9_MOUSE
|
||||||
| NC score | 0.036595 (rank : 69) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C7M3, Q6ZQE5, Q80WT6, Q8C7Z4, Q8CC32, Q8CEG2, Q99PQ3 | Gene names | Trim9, Kiaa0282 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9. | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.036587 (rank : 70) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.034100 (rank : 71) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.033149 (rank : 72) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
INHA_HUMAN
|
||||||
| NC score | 0.033060 (rank : 73) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05111 | Gene names | INHA | |||
|
Domain Architecture |

|
|||||
| Description | Inhibin alpha chain precursor. | |||||
|
CLUA1_HUMAN
|
||||||
| NC score | 0.032627 (rank : 74) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96AJ1, O75138, Q65ZA3, Q9H8R4, Q9H8T1 | Gene names | CLUAP1, KIAA0643 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.032506 (rank : 75) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
CLUA1_MOUSE
|
||||||
| NC score | 0.032302 (rank : 76) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
IPO9_HUMAN
|
||||||
| NC score | 0.027979 (rank : 77) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96P70, Q8N1Y1, Q8N3I2, Q8NCG9, Q96SU6, Q9NW01, Q9P0A8, Q9ULM8 | Gene names | IPO9, IMP9, KIAA1192, RANBP9 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-9 (Imp9) (Ran-binding protein 9) (RanbP9). | |||||
|
IPO9_MOUSE
|
||||||
| NC score | 0.027916 (rank : 78) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE6, Q924M3, Q924M4 | Gene names | Ipo9, Imp9, Ranbp9 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-9 (Importin 9a) (Imp9a) (Importin 9b) (Imp9b) (Ran-binding protein 9) (RanBP9). | |||||
|
RN122_HUMAN
|
||||||
| NC score | 0.026557 (rank : 79) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H9V4, Q52LK3 | Gene names | RNF122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
PPR3D_HUMAN
|
||||||
| NC score | 0.026264 (rank : 80) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95685, Q6DK02 | Gene names | PPP1R3D, PPP1R6 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 3D (Protein phosphatase 1, regulatory subunit 6) (Protein phosphatase 1-binding subunit R6). | |||||
|
RN141_HUMAN
|
||||||
| NC score | 0.025274 (rank : 81) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WVD5, Q9NZB4 | Gene names | RNF141, ZNF230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 141 (Zinc finger protein 230). | |||||
|
RNF6_HUMAN
|
||||||
| NC score | 0.025148 (rank : 82) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y252, Q9UF41 | Gene names | RNF6 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 6 (RING-H2 protein). | |||||
|
RN122_MOUSE
|
||||||
| NC score | 0.024303 (rank : 83) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BP31, Q80VA7, Q8BGD3 | Gene names | Rnf122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
RN141_MOUSE
|
||||||
| NC score | 0.023941 (rank : 84) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99MB7, Q9D040, Q9D5C4, Q9D9L0 | Gene names | Rnf141, Znf230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 141 (Zinc finger protein 230). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.023253 (rank : 85) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.022971 (rank : 86) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.021408 (rank : 87) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ARMC6_HUMAN
|
||||||
| NC score | 0.021050 (rank : 88) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.017827 (rank : 89) | θ value | 0.163984 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.017502 (rank : 90) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
THUM1_MOUSE
|
||||||
| NC score | 0.017396 (rank : 91) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99J36, Q3U6Y3 | Gene names | Thumpd1 | |||
|
Domain Architecture |

|
|||||
| Description | THUMP domain-containing protein 1. | |||||
|
PA2G3_HUMAN
|
||||||
| NC score | 0.016999 (rank : 92) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ20, O95768 | Gene names | PLA2G3 | |||
|
Domain Architecture |

|
|||||
| Description | Group 3 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group III secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GIII) (GIII sPLA2). | |||||
|
STX4_HUMAN
|
||||||
| NC score | 0.016402 (rank : 93) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12846, Q15525 | Gene names | STX4, STX4A | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-4 (NY-REN-31 antigen). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.016201 (rank : 94) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.015150 (rank : 95) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
CF049_HUMAN
|
||||||
| NC score | 0.015092 (rank : 96) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2TBC4, Q5T3D4, Q9NSV1 | Gene names | C6orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf49. | |||||
|
STX4_MOUSE
|
||||||
| NC score | 0.014659 (rank : 97) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70452 | Gene names | Stx4, Stx4a | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-4. | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.014529 (rank : 98) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.013940 (rank : 99) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
RTN2_MOUSE
|
||||||
| NC score | 0.013913 (rank : 100) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70622, O70620 | Gene names | Rtn2, Nspl1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.013683 (rank : 101) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.013627 (rank : 102) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.013402 (rank : 103) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
TF65_MOUSE
|
||||||
| NC score | 0.013195 (rank : 104) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
JIP4_HUMAN
|
||||||
| NC score | 0.013182 (rank : 105) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
ABLM1_MOUSE
|
||||||
| NC score | 0.013120 (rank : 106) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
ELL3_MOUSE
|
||||||
| NC score | 0.013053 (rank : 107) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80VR2 | Gene names | Ell3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
PYGO1_MOUSE
|
||||||
| NC score | 0.013007 (rank : 108) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0P5 | Gene names | Pygo1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
NBN_MOUSE
|
||||||
| NC score | 0.012802 (rank : 109) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.012794 (rank : 110) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.012782 (rank : 111) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
OST3B_MOUSE
|
||||||
| NC score | 0.012538 (rank : 112) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZS6 | Gene names | Hs3st3b1, 3ost3b1, Hs3st3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (m3-OST-3B). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.012267 (rank : 113) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
ADA18_MOUSE
|
||||||
| NC score | 0.011975 (rank : 114) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R157, Q60621 | Gene names | Adam18, Adam27, Dtgn3, Tmdc3 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 18 precursor (A disintegrin and metalloproteinase domain 18) (Transmembrane metalloproteinase-like, disintegrin-like, and cysteine- rich protein III) (tMDC III) (ADAM 27). | |||||
|
ASPH_HUMAN
|
||||||
| NC score | 0.011841 (rank : 115) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.011372 (rank : 116) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.011358 (rank : 117) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.011282 (rank : 118) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
NC6IP_HUMAN
|
||||||
| NC score | 0.011169 (rank : 119) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RS0, Q5GH23, Q8TDG9, Q96QU3, Q9H5V3 | Gene names | NCOA6IP, PIMT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT) (CLL-associated antigen KW-2) (Hepatocellular carcinoma-associated antigen 137) (HCA137). | |||||
|
AAKG3_MOUSE
|
||||||
| NC score | 0.011004 (rank : 120) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGM7, Q80WK8, Q8CJ41 | Gene names | Prkag3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-3 (AMPK gamma-3 chain) (AMPK gamma3). | |||||
|
B3A4_HUMAN
|
||||||
| NC score | 0.010983 (rank : 121) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96Q91, Q96RM5, Q9BXF2, Q9BXN3 | Gene names | SLC4A9, AE4, SBC5 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 4 (Anion exchanger 4) (Solute carrier family 4 member 9) (Sodium bicarbonate cotransporter 5). | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.010390 (rank : 122) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.010157 (rank : 123) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
EGFL7_HUMAN
|
||||||
| NC score | 0.010156 (rank : 124) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UHF1, Q5M7Y5, Q5VUD5, Q96EG0 | Gene names | EGFL7, MEGF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 7 precursor (Multiple EGF-like domain protein 7) (Multiple epidermal growth factor-like domain protein 7) (Vascular endothelial statin) (VE-statin) (NOTCH4-like protein) (ZNEU1). | |||||
|
PCSK5_HUMAN
|
||||||
| NC score | 0.009853 (rank : 125) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
CF182_MOUSE
|
||||||
| NC score | 0.009647 (rank : 126) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.009548 (rank : 127) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
MICA2_HUMAN
|
||||||
| NC score | 0.009456 (rank : 128) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.009120 (rank : 129) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
ARHGC_MOUSE
|
||||||
| NC score | 0.008537 (rank : 130) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.008497 (rank : 131) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.008250 (rank : 132) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
PLCB3_HUMAN
|
||||||
| NC score | 0.008062 (rank : 133) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.008016 (rank : 134) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.007839 (rank : 135) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CT077_HUMAN
|
||||||
| NC score | 0.007703 (rank : 136) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQG5 | Gene names | C20orf77 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf77. | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.007697 (rank : 137) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.007552 (rank : 138) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.007363 (rank : 139) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.006819 (rank : 140) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
PLXA4_HUMAN
|
||||||
| NC score | 0.006802 (rank : 141) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.006540 (rank : 142) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.005487 (rank : 143) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.005418 (rank : 144) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.005386 (rank : 145) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.005038 (rank : 146) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
ATS13_MOUSE
|
||||||
| NC score | 0.004566 (rank : 147) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
SYT14_MOUSE
|
||||||
| NC score | 0.004446 (rank : 148) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
PACN3_HUMAN
|
||||||
| NC score | 0.004217 (rank : 149) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
NFKB1_HUMAN
|
||||||
| NC score | 0.003824 (rank : 150) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19838, Q68D84, Q86V43, Q8N4X7, Q9NZC0 | Gene names | NFKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) [Contains: Nuclear factor NF-kappa-B p50 subunit]. | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.003766 (rank : 151) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.002980 (rank : 152) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.002710 (rank : 153) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.002471 (rank : 154) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.002397 (rank : 155) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.000347 (rank : 156) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
TMPS6_MOUSE
|
||||||
| NC score | 0.000107 (rank : 157) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.000102 (rank : 158) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
TAOK3_MOUSE
|
||||||
| NC score | -0.001888 (rank : 159) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
TAOK3_HUMAN
|
||||||
| NC score | -0.002035 (rank : 160) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1494 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H2K8, Q658N1, Q8IUM4, Q9HC79, Q9NZM9, Q9UHG7 | Gene names | TAOK3, DPK, JIK, KDS, MAP3K18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3) (Jun kinase-inhibitory kinase) (JNK/SAPK- inhibitory kinase) (Dendritic cell-derived protein kinase) (Cutaneous T-cell lymphoma tumor antigen HD-CL-09) (CTCL tumor antigen HD-CL-09) (Kinase from chicken homolog A) (hKFC-A). | |||||
|
IGF1R_HUMAN
|
||||||
| NC score | -0.002797 (rank : 161) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | -0.003790 (rank : 162) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
ZN425_HUMAN
|
||||||
| NC score | -0.003975 (rank : 163) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6IV72 | Gene names | ZNF425 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 425. | |||||