Please be patient as the page loads
|
HERC2_MOUSE
|
||||||
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HERC2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998295 (rank : 2) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 167 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HECD3_MOUSE
|
||||||
| θ value | 3.98324e-48 (rank : 3) | NC score | 0.707609 (rank : 5) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_HUMAN
|
||||||
| θ value | 1.15895e-47 (rank : 4) | NC score | 0.705717 (rank : 6) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HERC3_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 5) | NC score | 0.837281 (rank : 3) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
RCBT2_MOUSE
|
||||||
| θ value | 5.96599e-36 (rank : 6) | NC score | 0.528763 (rank : 44) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 4.27553e-34 (rank : 7) | NC score | 0.623115 (rank : 19) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
NEK8_HUMAN
|
||||||
| θ value | 5.584e-34 (rank : 8) | NC score | 0.153799 (rank : 67) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86SG6, Q2M1S6, Q8NDH1 | Gene names | NEK8, JCK, NEK12A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8) (NIMA-related kinase 12a). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 7.29293e-34 (rank : 9) | NC score | 0.627882 (rank : 15) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
RCBT2_HUMAN
|
||||||
| θ value | 3.61944e-33 (rank : 10) | NC score | 0.528991 (rank : 43) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
HERC5_HUMAN
|
||||||
| θ value | 1.79631e-32 (rank : 11) | NC score | 0.824295 (rank : 4) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 12) | NC score | 0.615190 (rank : 27) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 13) | NC score | 0.620034 (rank : 23) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 14) | NC score | 0.628144 (rank : 13) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
RCBT1_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 15) | NC score | 0.502828 (rank : 45) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 16) | NC score | 0.625012 (rank : 18) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 17) | NC score | 0.626794 (rank : 16) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
NEK9_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 18) | NC score | 0.159506 (rank : 66) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 19) | NC score | 0.620410 (rank : 22) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
NEK8_MOUSE
|
||||||
| θ value | 7.54701e-31 (rank : 20) | NC score | 0.153056 (rank : 68) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZR4, Q3U498, Q9D685 | Gene names | Nek8, Jck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8). | |||||
|
NEK9_MOUSE
|
||||||
| θ value | 7.54701e-31 (rank : 21) | NC score | 0.161483 (rank : 65) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 890 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K1R7, Q8R3P1 | Gene names | Nek9, Nercc | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9). | |||||
|
RCC2_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 22) | NC score | 0.621506 (rank : 21) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 23) | NC score | 0.629026 (rank : 12) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 24) | NC score | 0.626719 (rank : 17) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 6.38894e-30 (rank : 25) | NC score | 0.618082 (rank : 26) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
RCBT1_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 26) | NC score | 0.501400 (rank : 46) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
RCC2_MOUSE
|
||||||
| θ value | 3.17079e-29 (rank : 27) | NC score | 0.618815 (rank : 25) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 9.2256e-29 (rank : 28) | NC score | 0.574545 (rank : 35) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
SRGEF_MOUSE
|
||||||
| θ value | 9.2256e-29 (rank : 29) | NC score | 0.619476 (rank : 24) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
SRGEF_HUMAN
|
||||||
| θ value | 2.68423e-28 (rank : 30) | NC score | 0.622408 (rank : 20) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 3.50572e-28 (rank : 31) | NC score | 0.572544 (rank : 37) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 32) | NC score | 0.573166 (rank : 36) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 33) | NC score | 0.586502 (rank : 33) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 34) | NC score | 0.582219 (rank : 34) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
RCC1_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 35) | NC score | 0.610619 (rank : 28) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |
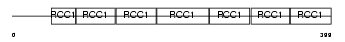
|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
RPGR_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 36) | NC score | 0.571323 (rank : 38) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 37) | NC score | 0.648142 (rank : 7) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
RCC1_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 38) | NC score | 0.597096 (rank : 32) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8VE37, Q3UDB6 | Gene names | Rcc1, Chc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 39) | NC score | 0.606313 (rank : 31) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 40) | NC score | 0.638624 (rank : 8) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 41) | NC score | 0.629561 (rank : 11) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 42) | NC score | 0.628126 (rank : 14) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 43) | NC score | 0.449229 (rank : 47) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 44) | NC score | 0.635328 (rank : 10) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
CUL7_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 45) | NC score | 0.322131 (rank : 52) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 46) | NC score | 0.635828 (rank : 9) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 47) | NC score | 0.254636 (rank : 54) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
WBS16_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 48) | NC score | 0.566496 (rank : 40) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CYF5 | Gene names | Wbscr16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein homolog. | |||||
|
PARC_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 49) | NC score | 0.260948 (rank : 53) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
WBS16_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 50) | NC score | 0.567925 (rank : 39) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I51, Q548B1, Q9H0G7 | Gene names | WBSCR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein (RCC1-like G exchanging factor-like protein). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 51) | NC score | 0.537304 (rank : 42) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 52) | NC score | 0.537684 (rank : 41) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 53) | NC score | 0.414944 (rank : 48) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 54) | NC score | 0.409578 (rank : 49) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
K0317_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 55) | NC score | 0.608232 (rank : 29) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 56) | NC score | 0.607444 (rank : 30) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
MIB2_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 57) | NC score | 0.114993 (rank : 85) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
MIB2_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 58) | NC score | 0.114709 (rank : 86) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 59) | NC score | 0.328250 (rank : 51) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 60) | NC score | 0.329008 (rank : 50) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MIB1_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 61) | NC score | 0.102245 (rank : 94) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MIB1_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 62) | NC score | 0.102397 (rank : 93) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
SQSTM_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 63) | NC score | 0.176672 (rank : 62) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
SQSTM_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 64) | NC score | 0.176242 (rank : 63) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
ZSWM2_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 65) | NC score | 0.139347 (rank : 77) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
ZSWM2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 66) | NC score | 0.126084 (rank : 80) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
CYB5_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 67) | NC score | 0.148704 (rank : 71) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P00167 | Gene names | CYB5 | |||
|
Domain Architecture |
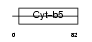
|
|||||
| Description | Cytochrome b5. | |||||
|
DTNA_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 68) | NC score | 0.111100 (rank : 89) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
CYB5B_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 69) | NC score | 0.140978 (rank : 75) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQX2, Q9D1M6, Q9D8R3 | Gene names | Cyb5b, Cyb5m | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome b5 type B precursor (Cytochrome b5 outer mitochondrial membrane isoform). | |||||
|
DTNA_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 70) | NC score | 0.113322 (rank : 87) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |
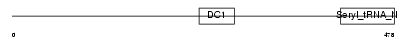
|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
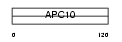
APC10_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 71) | NC score | 0.196244 (rank : 58) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UM13, Q2V500, Q9UG51, Q9Y5R0 | Gene names | ANAPC10, APC10 | |||
|
Domain Architecture |
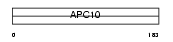
|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
CYB5B_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 72) | NC score | 0.133373 (rank : 78) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43169, Q96CC3, Q9BT35 | Gene names | CYB5B, CYB5M, OMB5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b5 type B precursor (Cytochrome b5 outer mitochondrial membrane isoform). | |||||
|
DTNB_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 73) | NC score | 0.108941 (rank : 90) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60941, O43782, O60881, O75538, Q9UE14, Q9UE15, Q9UE16 | Gene names | DTNB | |||
|
Domain Architecture |
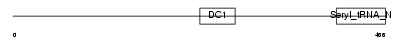
|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B). | |||||
|
APC10_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 74) | NC score | 0.191032 (rank : 59) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2H6, Q9CW24 | Gene names | Anapc10, Apc10 | |||
|
Domain Architecture |
|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
DTNB_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 75) | NC score | 0.105327 (rank : 92) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
FBX24_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 76) | NC score | 0.233178 (rank : 55) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75426, Q9H0G1 | Gene names | FBXO24, FBX24 | |||
|
Domain Architecture |
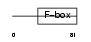
|
|||||
| Description | F-box only protein 24. | |||||
|
CYB5_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 77) | NC score | 0.125997 (rank : 81) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56395 | Gene names | Cyb5 | |||
|
Domain Architecture |
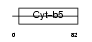
|
|||||
| Description | Cytochrome b5. | |||||
|
FBX24_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 78) | NC score | 0.209722 (rank : 56) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D417 | Gene names | Fbxo24, Fbx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 24. | |||||
|
DRP2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 79) | NC score | 0.107844 (rank : 91) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
MFGM_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 80) | NC score | 0.020414 (rank : 136) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |
|
|||||
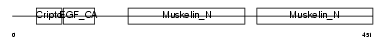
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
SAKS1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 81) | NC score | 0.057757 (rank : 107) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
SAKS1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 82) | NC score | 0.056553 (rank : 110) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q922Y1, Q3UCP8 | Gene names | Saks1, D19Ertd721e | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein) (mY33K) (Protein 2B28). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 83) | NC score | 0.046526 (rank : 115) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
NBR1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 84) | NC score | 0.080632 (rank : 102) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14596, Q13173, Q15026, Q96GB6, Q9NRF7 | Gene names | NBR1, 1A13B, KIAA0049, M17S2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2) (1A1-3B). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.163984 (rank : 85) | NC score | 0.026111 (rank : 127) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 86) | NC score | 0.046179 (rank : 116) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 87) | NC score | 0.042282 (rank : 118) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
LRFN4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 88) | NC score | 0.009669 (rank : 164) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PJG9, Q9BWJ0 | Gene names | LRFN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 4 precursor. | |||||
|
SPAG6_MOUSE
|
||||||
| θ value | 0.279714 (rank : 89) | NC score | 0.021862 (rank : 133) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
MAK_MOUSE
|
||||||
| θ value | 0.365318 (rank : 90) | NC score | 0.019798 (rank : 137) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04859 | Gene names | Mak, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MAK (EC 2.7.11.22) (Male germ cell- associated kinase) (Protein kinase RCK). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 0.62314 (rank : 91) | NC score | 0.044288 (rank : 117) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 0.62314 (rank : 92) | NC score | 0.040268 (rank : 121) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 93) | NC score | 0.020656 (rank : 135) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 0.62314 (rank : 94) | NC score | 0.042226 (rank : 119) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
CU056_HUMAN
|
||||||
| θ value | 0.813845 (rank : 95) | NC score | 0.024432 (rank : 130) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0A9, Q9NSE5 | Gene names | C21orf56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56. | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 96) | NC score | 0.028981 (rank : 124) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 97) | NC score | 0.012574 (rank : 160) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 98) | NC score | 0.018422 (rank : 142) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
XRCC4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 99) | NC score | 0.035520 (rank : 123) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q924T3, Q8BKC9, Q8BU02, Q9D6U6 | Gene names | Xrcc4 | |||
|
Domain Architecture |
|
|||||
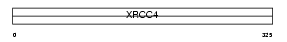
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
ZNF85_HUMAN
|
||||||
| θ value | 1.06291 (rank : 100) | NC score | -0.003688 (rank : 194) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03923 | Gene names | ZNF85 | |||
|
Domain Architecture |
|
|||||
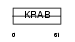
| Description | Zinc finger protein 85 (Zinc finger protein HPF4) (HTF1). | |||||
|
ABCAD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 101) | NC score | 0.010115 (rank : 163) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 102) | NC score | 0.017483 (rank : 146) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 103) | NC score | 0.022607 (rank : 132) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
HEMK1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 104) | NC score | 0.040663 (rank : 120) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921L7 | Gene names | Hemk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HemK methyltransferase family member 1 (EC 2.1.1.-). | |||||
|
ITA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 105) | NC score | 0.006960 (rank : 170) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13612 | Gene names | ITGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (CD49d antigen). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 106) | NC score | 0.012778 (rank : 159) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MFN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 107) | NC score | 0.018640 (rank : 141) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U63, Q3V3B8, Q80WP4, Q80XK3, Q8BHF0, Q8BKV5, Q8R535, Q923X2 | Gene names | Mfn2, Kiaa0214, Marf | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN2 (EC 3.6.5.-) (Mitofusin-2) (Hypertension- related protein 1) (Mitochondrial assembly regulatory factor) (HSG protein). | |||||
|
PDZD8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 108) | NC score | 0.024418 (rank : 131) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 109) | NC score | 0.025609 (rank : 129) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 110) | NC score | 0.025954 (rank : 128) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 111) | NC score | 0.017578 (rank : 145) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
NBR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 112) | NC score | 0.058811 (rank : 106) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
SUOX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 113) | NC score | 0.047992 (rank : 114) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51687 | Gene names | SUOX | |||
|
Domain Architecture |

|
|||||
| Description | Sulfite oxidase, mitochondrial precursor (EC 1.8.3.1). | |||||
|
TOP2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 114) | NC score | 0.018868 (rank : 140) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
KPCE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 115) | NC score | 0.016011 (rank : 151) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q02156, Q53SL4, Q53SM5, Q9UE81 | Gene names | PRKCE, PKCE | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
ADAM9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.003394 (rank : 187) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61072, Q60618, Q61853, Q80U94 | Gene names | Adam9, Kiaa0021, Mdc9, Mltng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma). | |||||
|
K1377_HUMAN
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.014777 (rank : 156) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2H0, Q4G0U6 | Gene names | KIAA1377 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1377. | |||||
|
PRDM4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | -0.004521 (rank : 195) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
WDR67_HUMAN
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | 0.006267 (rank : 173) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
DICER_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.015415 (rank : 152) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.015132 (rank : 153) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
NNTM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.017584 (rank : 144) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61941, Q9JK70 | Gene names | Nnt | |||
|
Domain Architecture |

|
|||||
| Description | NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase). | |||||
|
SLAF7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.016280 (rank : 150) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQ25, Q8N6Y8, Q8ND32, Q9NY08, Q9NY23 | Gene names | SLAMF7, CS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLAM family member 7 precursor (CD2-like receptor activating cytotoxic cells) (CRACC) (Protein 19A) (CD2 subset 1) (Novel Ly9) (Membrane protein FOAP-12) (CD319 antigen). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.016544 (rank : 149) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.016879 (rank : 148) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
TAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.002681 (rank : 192) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36371 | Gene names | Tap2, Abcb3, Ham-2, Ham2 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen peptide transporter 2 (APT2) (ATP-binding cassette sub-family B member 3) (Histocompatibility antigen modifier 2). | |||||
|
GP110_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.006605 (rank : 171) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEC3, Q8BMV8 | Gene names | Gpr110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 110 precursor. | |||||
|
HIPK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.017011 (rank : 147) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86Z02, O75125, Q8IYD7, Q8NDN5, Q8NEB6, Q8TBZ1 | Gene names | HIPK1, KIAA0630 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1). | |||||
|
IMMT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.007161 (rank : 169) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CAQ8, Q66JS4, Q7TNE2, Q8C7V1, Q8CCI0, Q8CDA8, Q9D9F6 | Gene names | Immt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin). | |||||
|
JHD1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.003984 (rank : 185) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHM5, Q8NCI2, Q96HC7, Q96SL0, Q96T03, Q9NS96, Q9UF75 | Gene names | FBXL10, CXXC2, FBL10, JHDM1B, PCCX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10) (Protein JEMMA) (Jumonji domain-containing EMSY-interactor methyltransferase motif protein) (CXXC-type zinc finger protein 2) (Protein-containing CXXC domain 2). | |||||
|
NTRI_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.005039 (rank : 179) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PJ0 | Gene names | Nt, Hnt | |||
|
Domain Architecture |
|
|||||
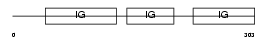
| Description | Neurotrimin precursor. | |||||
|
SETD7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.019287 (rank : 139) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
STK38_MOUSE
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.015061 (rank : 154) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VJ4 | Gene names | Stk38, Ndr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 38 (EC 2.7.11.1) (NDR1 protein kinase) (Nuclear Dbf2-related kinase 1). | |||||
|
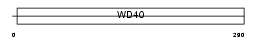
STRAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.005765 (rank : 175) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3F4, Q9UQC8 | Gene names | STRAP, MAWD, UNRIP | |||
|
Domain Architecture |
|
|||||
| Description | Serine-threonine kinase receptor-associated protein (UNR-interacting protein) (WD-40 repeat protein PT-WD) (MAP activator with WD repeats). | |||||
|
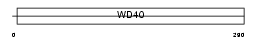
STRAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.005623 (rank : 176) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1Z2 | Gene names | Strap, Unrip | |||
|
Domain Architecture |
|
|||||
| Description | Serine-threonine kinase receptor-associated protein (UNR-interacting protein). | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.013623 (rank : 157) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
ZN431_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | -0.004958 (rank : 196) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF32, Q8IWC4 | Gene names | ZNF431, KIAA1969 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 431. | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.019579 (rank : 138) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BEGIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.005833 (rank : 174) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
CATZ_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.004808 (rank : 183) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUU7 | Gene names | Ctsz | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-). | |||||
|
CN046_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.027831 (rank : 125) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q52LA3 | Gene names | C14orf46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf46. | |||||
|
CN046_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.027831 (rank : 126) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CD94 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf46 homolog. | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.021636 (rank : 134) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
HSH2D_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.011357 (rank : 162) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
LRC25_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.018271 (rank : 143) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N386, Q8N9A5 | Gene names | LRRC25, MAPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 25 precursor (Monocyte and plasmacytoid-activated protein). | |||||
|
MFN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.011371 (rank : 161) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95140, O95572, Q5JXC3, Q5JXC4, Q9H131, Q9NSX8 | Gene names | MFN2, CPRP1, KIAA0214 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN2 (EC 3.6.5.-) (Mitofusin-2). | |||||
|
NTRI_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.004336 (rank : 184) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P121, Q6UXJ3, Q86VJ9 | Gene names | NT | |||
|
Domain Architecture |
|
|||||
| Description | Neurotrimin precursor (hNT). | |||||
|
NXF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.005372 (rank : 177) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBU9, Q99799, Q9UQL2 | Gene names | NXF1, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear RNA export factor 1 (Tip-associating protein) (Tip-associated protein) (mRNA export factor TAP). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.003185 (rank : 190) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
STK38_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.014853 (rank : 155) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15208 | Gene names | STK38, NDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 38 (EC 2.7.11.1) (NDR1 protein kinase) (Nuclear Dbf2-related kinase 1). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.013017 (rank : 158) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
U3IP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.003339 (rank : 188) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43818, Q8IZ30 | Gene names | RNU3IP2, U355K | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar RNA-interacting protein 2 (U3 small nucleolar ribonucleoprotein-associated 55 kDa protein) (U3 snoRNP-associated 55 kDa protein) (U3-55K). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.003082 (rank : 191) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
CSAD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.008362 (rank : 168) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y600, Q9UNJ5, Q9Y601 | Gene names | CSAD, CSD | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine sulfinic acid decarboxylase (EC 4.1.1.29) (Sulfinoalanine decarboxylase) (Cysteine-sulfinate decarboxylase). | |||||
|
CSAD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.008405 (rank : 167) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBE0 | Gene names | Csad | |||
|
Domain Architecture |
|
|||||
| Description | Cysteine sulfinic acid decarboxylase (EC 4.1.1.29) (Sulfinoalanine decarboxylase) (Cysteine-sulfinate decarboxylase). | |||||
|
DUS4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.004916 (rank : 181) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.004953 (rank : 180) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.003552 (rank : 186) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
ITA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.004873 (rank : 182) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08648, Q96HA5 | Gene names | ITGA5, FNRA | |||
|
Domain Architecture |
|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
PK1IP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.003228 (rank : 189) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DCE5, Q3UBK7, Q80UT4, Q8C5N6, Q923K2, Q9D3E9 | Gene names | Pak1ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p21-activated protein kinase-interacting protein 1 (PAK1-interacting protein 1) (Putative PAK inhibitor Skb15). | |||||
|
PLCB4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | -0.000025 (rank : 193) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
PRDM4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | -0.005122 (rank : 197) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
SGOL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.005165 (rank : 178) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
SUOX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.037502 (rank : 122) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R086 | Gene names | Suox | |||
|
Domain Architecture |

|
|||||
| Description | Sulfite oxidase, mitochondrial precursor (EC 1.8.3.1). | |||||
|
SYLC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.008574 (rank : 166) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2J5, Q9NSE1 | Gene names | LARS, KIAA1352 | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.008794 (rank : 165) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMJ2, Q8BKW9 | Gene names | Lars | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.006399 (rank : 172) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
ABTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054946 (rank : 113) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ABTB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.057313 (rank : 109) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
APBB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.059569 (rank : 105) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.057424 (rank : 108) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
BAG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.132646 (rank : 79) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.112045 (rank : 88) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
GAS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.088568 (rank : 100) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
GAS7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.088490 (rank : 101) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.092969 (rank : 99) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.093324 (rank : 98) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.093502 (rank : 97) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.100462 (rank : 95) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
PIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.142501 (rank : 72) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PIN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.141562 (rank : 73) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PINL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.169691 (rank : 64) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.099189 (rank : 96) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.075690 (rank : 104) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.076795 (rank : 103) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
SAV1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.181762 (rank : 61) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
SAV1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.181978 (rank : 60) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
STXB4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.055516 (rank : 111) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
STXB4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.055025 (rank : 112) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
WWC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.150615 (rank : 70) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.151408 (rank : 69) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
WWOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.141173 (rank : 74) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
WWOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.139913 (rank : 76) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
WWTR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.120953 (rank : 83) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.123484 (rank : 82) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.118112 (rank : 84) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
YAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.201905 (rank : 57) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 167 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.998295 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.837281 (rank : 3) | θ value | 1.73182e-43 (rank : 5) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.824295 (rank : 4) | θ value | 1.79631e-32 (rank : 11) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.707609 (rank : 5) | θ value | 3.98324e-48 (rank : 3) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.705717 (rank : 6) | θ value | 1.15895e-47 (rank : 4) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.648142 (rank : 7) | θ value | 2.12685e-25 (rank : 37) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.638624 (rank : 8) | θ value | 4.15078e-21 (rank : 40) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.635828 (rank : 9) | θ value | 2.5182e-18 (rank : 46) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.635328 (rank : 10) | θ value | 8.65492e-19 (rank : 44) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.629561 (rank : 11) | θ value | 2.27762e-19 (rank : 41) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.629026 (rank : 12) | θ value | 4.89182e-30 (rank : 23) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.628144 (rank : 13) | θ value | 8.91499e-32 (rank : 14) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.628126 (rank : 14) | θ value | 6.62687e-19 (rank : 42) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.627882 (rank : 15) | θ value | 7.29293e-34 (rank : 9) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.626794 (rank : 16) | θ value | 1.52067e-31 (rank : 17) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.626719 (rank : 17) | θ value | 4.89182e-30 (rank : 24) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.625012 (rank : 18) | θ value | 1.52067e-31 (rank : 16) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.623115 (rank : 19) | θ value | 4.27553e-34 (rank : 7) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
SRGEF_HUMAN
|
||||||
| NC score | 0.622408 (rank : 20) | θ value | 2.68423e-28 (rank : 30) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
RCC2_HUMAN
|
||||||
| NC score | 0.621506 (rank : 21) | θ value | 2.19584e-30 (rank : 22) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.620410 (rank : 22) | θ value | 5.77852e-31 (rank : 19) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.620034 (rank : 23) | θ value | 5.22648e-32 (rank : 13) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
SRGEF_MOUSE
|
||||||
| NC score | 0.619476 (rank : 24) | θ value | 9.2256e-29 (rank : 29) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
RCC2_MOUSE
|
||||||
| NC score | 0.618815 (rank : 25) | θ value | 3.17079e-29 (rank : 27) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.618082 (rank : 26) | θ value | 6.38894e-30 (rank : 25) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.615190 (rank : 27) | θ value | 5.22648e-32 (rank : 12) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
RCC1_HUMAN
|
||||||
| NC score | 0.610619 (rank : 28) | θ value | 5.06226e-27 (rank : 35) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
K0317_HUMAN
|
||||||
| NC score | 0.608232 (rank : 29) | θ value | 7.58209e-15 (rank : 55) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| NC score | 0.607444 (rank : 30) | θ value | 1.29331e-14 (rank : 56) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.606313 (rank : 31) | θ value | 8.36355e-22 (rank : 39) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
RCC1_MOUSE
|
||||||
| NC score | 0.597096 (rank : 32) | θ value | 3.75424e-22 (rank : 38) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8VE37, Q3UDB6 | Gene names | Rcc1, Chc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.586502 (rank : 33) | θ value | 7.80994e-28 (rank : 33) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.582219 (rank : 34) | θ value | 1.33217e-27 (rank : 34) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.574545 (rank : 35) | θ value | 9.2256e-29 (rank : 28) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.573166 (rank : 36) | θ value | 3.50572e-28 (rank : 32) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.572544 (rank : 37) | θ value | 3.50572e-28 (rank : 31) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
RPGR_HUMAN
|
||||||
| NC score | 0.571323 (rank : 38) | θ value | 1.12775e-26 (rank : 36) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
WBS16_HUMAN
|
||||||
| NC score | 0.567925 (rank : 39) | θ value | 1.24977e-17 (rank : 50) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I51, Q548B1, Q9H0G7 | Gene names | WBSCR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein (RCC1-like G exchanging factor-like protein). | |||||
|
WBS16_MOUSE
|
||||||
| NC score | 0.566496 (rank : 40) | θ value | 5.60996e-18 (rank : 48) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CYF5 | Gene names | Wbscr16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein homolog. | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.537684 (rank : 41) | θ value | 5.25075e-16 (rank : 52) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.537304 (rank : 42) | θ value | 5.25075e-16 (rank : 51) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
RCBT2_HUMAN
|
||||||
| NC score | 0.528991 (rank : 43) | θ value | 3.61944e-33 (rank : 10) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
RCBT2_MOUSE
|
||||||
| NC score | 0.528763 (rank : 44) | θ value | 5.96599e-36 (rank : 6) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
RCBT1_MOUSE
|
||||||
| NC score | 0.502828 (rank : 45) | θ value | 1.16434e-31 (rank : 15) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
RCBT1_HUMAN
|
||||||
| NC score | 0.501400 (rank : 46) | θ value | 2.42779e-29 (rank : 26) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.449229 (rank : 47) | θ value | 8.65492e-19 (rank : 43) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.414944 (rank : 48) | θ value | 8.95645e-16 (rank : 53) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.409578 (rank : 49) | θ value | 1.52774e-15 (rank : 54) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.329008 (rank : 50) | θ value | 2.52405e-10 (rank : 60) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.328250 (rank : 51) | θ value | 1.47974e-10 (rank : 59) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.322131 (rank : 52) | θ value | 1.13037e-18 (rank : 45) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
PARC_MOUSE
|
||||||
| NC score | 0.260948 (rank : 53) | θ value | 1.24977e-17 (rank : 49) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.254636 (rank : 54) | θ value | 5.60996e-18 (rank : 47) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
FBX24_HUMAN
|
||||||
| NC score | 0.233178 (rank : 55) | θ value | 0.0113563 (rank : 76) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75426, Q9H0G1 | Gene names | FBXO24, FBX24 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 24. | |||||
|
FBX24_MOUSE
|
||||||
| NC score | 0.209722 (rank : 56) | θ value | 0.0252991 (rank : 78) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D417 | Gene names | Fbxo24, Fbx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 24. | |||||
|
YAP1_MOUSE
|
||||||
| NC score | 0.201905 (rank : 57) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
APC10_HUMAN
|
||||||
| NC score | 0.196244 (rank : 58) | θ value | 0.00509761 (rank : 71) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UM13, Q2V500, Q9UG51, Q9Y5R0 | Gene names | ANAPC10, APC10 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
APC10_MOUSE
|
||||||
| NC score | 0.191032 (rank : 59) | θ value | 0.00869519 (rank : 74) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2H6, Q9CW24 | Gene names | Anapc10, Apc10 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
SAV1_MOUSE
|
||||||
| NC score | 0.181978 (rank : 60) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
SAV1_HUMAN
|
||||||
| NC score | 0.181762 (rank : 61) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
SQSTM_MOUSE
|
||||||
| NC score | 0.176672 (rank : 62) | θ value | 3.19293e-05 (rank : 63) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
SQSTM_HUMAN
|
||||||
| NC score | 0.176242 (rank : 63) | θ value | 4.1701e-05 (rank : 64) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
PINL_HUMAN
|
||||||
| NC score | 0.169691 (rank : 64) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
NEK9_MOUSE
|
||||||
| NC score | 0.161483 (rank : 65) | θ value | 7.54701e-31 (rank : 21) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 890 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K1R7, Q8R3P1 | Gene names | Nek9, Nercc | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9). | |||||
|
NEK9_HUMAN
|
||||||
| NC score | 0.159506 (rank : 66) | θ value | 4.42448e-31 (rank : 18) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||
|
NEK8_HUMAN
|
||||||
| NC score | 0.153799 (rank : 67) | θ value | 5.584e-34 (rank : 8) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86SG6, Q2M1S6, Q8NDH1 | Gene names | NEK8, JCK, NEK12A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8) (NIMA-related kinase 12a). | |||||
|
NEK8_MOUSE
|
||||||
| NC score | 0.153056 (rank : 68) | θ value | 7.54701e-31 (rank : 20) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZR4, Q3U498, Q9D685 | Gene names | Nek8, Jck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8). | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.151408 (rank : 69) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
WWC1_HUMAN
|
||||||
| NC score | 0.150615 (rank : 70) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
CYB5_HUMAN
|
||||||
| NC score | 0.148704 (rank : 71) | θ value | 0.000786445 (rank : 67) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P00167 | Gene names | CYB5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b5. | |||||
|
PIN1_HUMAN
|
||||||
| NC score | 0.142501 (rank : 72) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PIN1_MOUSE
|
||||||
| NC score | 0.141562 (rank : 73) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
WWOX_HUMAN
|
||||||
| NC score | 0.141173 (rank : 74) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
CYB5B_MOUSE
|
||||||
| NC score | 0.140978 (rank : 75) | θ value | 0.00175202 (rank : 69) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQX2, Q9D1M6, Q9D8R3 | Gene names | Cyb5b, Cyb5m | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome b5 type B precursor (Cytochrome b5 outer mitochondrial membrane isoform). | |||||
|
WWOX_MOUSE
|
||||||
| NC score | 0.139913 (rank : 76) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
ZSWM2_HUMAN
|
||||||
| NC score | 0.139347 (rank : 77) | θ value | 4.1701e-05 (rank : 65) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
CYB5B_HUMAN
|
||||||
| NC score | 0.133373 (rank : 78) | θ value | 0.00509761 (rank : 72) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43169, Q96CC3, Q9BT35 | Gene names | CYB5B, CYB5M, OMB5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b5 type B precursor (Cytochrome b5 outer mitochondrial membrane isoform). | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.132646 (rank : 79) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
ZSWM2_MOUSE
|
||||||
| NC score | 0.126084 (rank : 80) | θ value | 0.000461057 (rank : 66) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
CYB5_MOUSE
|
||||||
| NC score | 0.125997 (rank : 81) | θ value | 0.0148317 (rank : 77) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56395 | Gene names | Cyb5 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b5. | |||||
|
WWTR1_MOUSE
|
||||||
| NC score | 0.123484 (rank : 82) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_HUMAN
|
||||||
| NC score | 0.120953 (rank : 83) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.118112 (rank : 84) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.114993 (rank : 85) | θ value | 3.52202e-12 (rank : 57) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
MIB2_MOUSE
|
||||||
| NC score | 0.114709 (rank : 86) | θ value | 8.67504e-11 (rank : 58) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
DTNA_MOUSE
|
||||||
| NC score | 0.113322 (rank : 87) | θ value | 0.00175202 (rank : 70) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.112045 (rank : 88) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
DTNA_HUMAN
|
||||||
| NC score | 0.111100 (rank : 89) | θ value | 0.00134147 (rank : 68) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
DTNB_HUMAN
|
||||||
| NC score | 0.108941 (rank : 90) | θ value | 0.00665767 (rank : 73) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60941, O43782, O60881, O75538, Q9UE14, Q9UE15, Q9UE16 | Gene names | DTNB | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B). | |||||
|
DRP2_HUMAN
|
||||||
| NC score | 0.107844 (rank : 91) | θ value | 0.0330416 (rank : 79) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
DTNB_MOUSE
|
||||||
| NC score | 0.105327 (rank : 92) | θ value | 0.00869519 (rank : 75) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
MIB1_MOUSE
|
||||||
| NC score | 0.102397 (rank : 93) | θ value | 3.29651e-10 (rank : 62) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
MIB1_HUMAN
|
||||||
| NC score | 0.102245 (rank : 94) | θ value | 3.29651e-10 (rank : 61) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.100462 (rank : 95) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.099189 (rank : 96) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.093502 (rank : 97) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.093324 (rank : 98) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.092969 (rank : 99) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
GAS7_HUMAN
|
||||||
| NC score | 0.088568 (rank : 100) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
GAS7_MOUSE
|
||||||
| NC score | 0.088490 (rank : 101) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
NBR1_HUMAN
|
||||||
| NC score | 0.080632 (rank : 102) | θ value | 0.0961366 (rank : 84) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14596, Q13173, Q15026, Q96GB6, Q9NRF7 | Gene names | NBR1, 1A13B, KIAA0049, M17S2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2) (1A1-3B). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.076795 (rank : 103) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.075690 (rank : 104) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.059569 (rank : 105) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
NBR1_MOUSE
|
||||||
| NC score | 0.058811 (rank : 106) | θ value | 1.81305 (rank : 112) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
SAKS1_HUMAN
|
||||||
| NC score | 0.057757 (rank : 107) | θ value | 0.0736092 (rank : 81) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.057424 (rank : 108) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
ABTB2_MOUSE
|
||||||
| NC score | 0.057313 (rank : 109) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
SAKS1_MOUSE
|
||||||
| NC score | 0.056553 (rank : 110) | θ value | 0.0736092 (rank : 82) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q922Y1, Q3UCP8 | Gene names | Saks1, D19Ertd721e | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein) (mY33K) (Protein 2B28). | |||||
|
STXB4_HUMAN
|
||||||
| NC score | 0.055516 (rank : 111) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
STXB4_MOUSE
|
||||||
| NC score | 0.055025 (rank : 112) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
ABTB2_HUMAN
|
||||||
| NC score | 0.054946 (rank : 113) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
SUOX_HUMAN
|
||||||
| NC score | 0.047992 (rank : 114) | θ value | 1.81305 (rank : 113) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51687 | Gene names | SUOX | |||
|
Domain Architecture |

|
|||||
| Description | Sulfite oxidase, mitochondrial precursor (EC 1.8.3.1). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.046526 (rank : 115) | θ value | 0.0961366 (rank : 83) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.046179 (rank : 116) | θ value | 0.279714 (rank : 86) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.044288 (rank : 117) | θ value | 0.62314 (rank : 91) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.042282 (rank : 118) | θ value | 0.279714 (rank : 87) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.042226 (rank : 119) | θ value | 0.62314 (rank : 94) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
HEMK1_MOUSE
|
||||||
| NC score | 0.040663 (rank : 120) | θ value | 1.38821 (rank : 104) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921L7 | Gene names | Hemk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HemK methyltransferase family member 1 (EC 2.1.1.-). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.040268 (rank : 121) | θ value | 0.62314 (rank : 92) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SUOX_MOUSE
|
||||||
| NC score | 0.037502 (rank : 122) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R086 | Gene names | Suox | |||
|
Domain Architecture |

|
|||||
| Description | Sulfite oxidase, mitochondrial precursor (EC 1.8.3.1). | |||||
|
XRCC4_MOUSE
|
||||||
| NC score | 0.035520 (rank : 123) | θ value | 1.06291 (rank : 99) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q924T3, Q8BKC9, Q8BU02, Q9D6U6 | Gene names | Xrcc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.028981 (rank : 124) | θ value | 1.06291 (rank : 96) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CN046_HUMAN
|
||||||
| NC score | 0.027831 (rank : 125) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q52LA3 | Gene names | C14orf46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf46. | |||||
|
CN046_MOUSE
|
||||||
| NC score | 0.027831 (rank : 126) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CD94 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf46 homolog. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.026111 (rank : 127) | θ value | 0.163984 (rank : 85) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.025954 (rank : 128) | θ value | 1.38821 (rank : 110) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.025609 (rank : 129) | θ value | 1.38821 (rank : 109) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
CU056_HUMAN
|
||||||
| NC score | 0.024432 (rank : 130) | θ value | 0.813845 (rank : 95) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0A9, Q9NSE5 | Gene names | C21orf56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56. | |||||
|
PDZD8_HUMAN
|
||||||
| NC score | 0.024418 (rank : 131) | θ value | 1.38821 (rank : 108) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.022607 (rank : 132) | θ value | 1.38821 (rank : 103) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
SPAG6_MOUSE
|
||||||
| NC score | 0.021862 (rank : 133) | θ value | 0.279714 (rank : 89) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.021636 (rank : 134) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.020656 (rank : 135) | θ value | 0.62314 (rank : 93) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
MFGM_MOUSE
|
||||||
| NC score | 0.020414 (rank : 136) | θ value | 0.0736092 (rank : 80) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
MAK_MOUSE
|
||||||
| NC score | 0.019798 (rank : 137) | θ value | 0.365318 (rank : 90) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04859 | Gene names | Mak, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MAK (EC 2.7.11.22) (Male germ cell- associated kinase) (Protein kinase RCK). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.019579 (rank : 138) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
SETD7_MOUSE
|
||||||
| NC score | 0.019287 (rank : 139) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
TOP2A_HUMAN
|
||||||
| NC score | 0.018868 (rank : 140) | θ value | 1.81305 (rank : 114) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
MFN2_MOUSE
|
||||||
| NC score | 0.018640 (rank : 141) | θ value | 1.38821 (rank : 107) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U63, Q3V3B8, Q80WP4, Q80XK3, Q8BHF0, Q8BKV5, Q8R535, Q923X2 | Gene names | Mfn2, Kiaa0214, Marf | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN2 (EC 3.6.5.-) (Mitofusin-2) (Hypertension- related protein 1) (Mitochondrial assembly regulatory factor) (HSG protein). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.018422 (rank : 142) | θ value | 1.06291 (rank : 98) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
LRC25_HUMAN
|
||||||
| NC score | 0.018271 (rank : 143) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N386, Q8N9A5 | Gene names | LRRC25, MAPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 25 precursor (Monocyte and plasmacytoid-activated protein). | |||||
|
NNTM_MOUSE
|
||||||
| NC score | 0.017584 (rank : 144) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61941, Q9JK70 | Gene names | Nnt | |||
|
Domain Architecture |

|
|||||
| Description | NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.017578 (rank : 145) | θ value | 1.81305 (rank : 111) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.017483 (rank : 146) | θ value | 1.38821 (rank : 102) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
HIPK1_HUMAN
|
||||||
| NC score | 0.017011 (rank : 147) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86Z02, O75125, Q8IYD7, Q8NDN5, Q8NEB6, Q8TBZ1 | Gene names | HIPK1, KIAA0630 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.016879 (rank : 148) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.016544 (rank : 149) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SLAF7_HUMAN
|
||||||
| NC score | 0.016280 (rank : 150) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQ25, Q8N6Y8, Q8ND32, Q9NY08, Q9NY23 | Gene names | SLAMF7, CS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLAM family member 7 precursor (CD2-like receptor activating cytotoxic cells) (CRACC) (Protein 19A) (CD2 subset 1) (Novel Ly9) (Membrane protein FOAP-12) (CD319 antigen). | |||||
|
KPCE_HUMAN
|
||||||
| NC score | 0.016011 (rank : 151) | θ value | 2.36792 (rank : 115) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q02156, Q53SL4, Q53SM5, Q9UE81 | Gene names | PRKCE, PKCE | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.015415 (rank : 152) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.015132 (rank : 153) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
STK38_MOUSE
|
||||||
| NC score | 0.015061 (rank : 154) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VJ4 | Gene names | Stk38, Ndr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 38 (EC 2.7.11.1) (NDR1 protein kinase) (Nuclear Dbf2-related kinase 1). | |||||
|
STK38_HUMAN
|
||||||
| NC score | 0.014853 (rank : 155) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15208 | Gene names | STK38, NDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 38 (EC 2.7.11.1) (NDR1 protein kinase) (Nuclear Dbf2-related kinase 1). | |||||
|
K1377_HUMAN
|
||||||
| NC score | 0.014777 (rank : 156) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2H0, Q4G0U6 | Gene names | KIAA1377 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1377. | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.013623 (rank : 157) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.013017 (rank : 158) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.012778 (rank : 159) | θ value | 1.38821 (rank : 106) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.012574 (rank : 160) | θ value | 1.06291 (rank : 97) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
MFN2_HUMAN
|
||||||
| NC score | 0.011371 (rank : 161) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95140, O95572, Q5JXC3, Q5JXC4, Q9H131, Q9NSX8 | Gene names | MFN2, CPRP1, KIAA0214 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN2 (EC 3.6.5.-) (Mitofusin-2). | |||||
|
HSH2D_MOUSE
|
||||||
| NC score | 0.011357 (rank : 162) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
ABCAD_HUMAN
|
||||||
| NC score | 0.010115 (rank : 163) | θ value | 1.38821 (rank : 101) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
LRFN4_HUMAN
|
||||||
| NC score | 0.009669 (rank : 164) | θ value | 0.279714 (rank : 88) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PJG9, Q9BWJ0 | Gene names | LRFN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 4 precursor. | |||||
|
SYLC_MOUSE
|
||||||
| NC score | 0.008794 (rank : 165) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMJ2, Q8BKW9 | Gene names | Lars | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLC_HUMAN
|
||||||
| NC score | 0.008574 (rank : 166) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2J5, Q9NSE1 | Gene names | LARS, KIAA1352 | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
CSAD_MOUSE
|
||||||
| NC score | 0.008405 (rank : 167) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBE0 | Gene names | Csad | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine sulfinic acid decarboxylase (EC 4.1.1.29) (Sulfinoalanine decarboxylase) (Cysteine-sulfinate decarboxylase). | |||||
|
CSAD_HUMAN
|
||||||
| NC score | 0.008362 (rank : 168) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y600, Q9UNJ5, Q9Y601 | Gene names | CSAD, CSD | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine sulfinic acid decarboxylase (EC 4.1.1.29) (Sulfinoalanine decarboxylase) (Cysteine-sulfinate decarboxylase). | |||||
|
IMMT_MOUSE
|
||||||
| NC score | 0.007161 (rank : 169) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CAQ8, Q66JS4, Q7TNE2, Q8C7V1, Q8CCI0, Q8CDA8, Q9D9F6 | Gene names | Immt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin). | |||||
|
ITA4_HUMAN
|
||||||
| NC score | 0.006960 (rank : 170) | θ value | 1.38821 (rank : 105) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13612 | Gene names | ITGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (CD49d antigen). | |||||
|
GP110_MOUSE
|
||||||
| NC score | 0.006605 (rank : 171) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEC3, Q8BMV8 | Gene names | Gpr110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 110 precursor. | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.006399 (rank : 172) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
WDR67_HUMAN
|
||||||
| NC score | 0.006267 (rank : 173) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
BEGIN_HUMAN
|
||||||
| NC score | 0.005833 (rank : 174) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
STRAP_HUMAN
|
||||||
| NC score | 0.005765 (rank : 175) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3F4, Q9UQC8 | Gene names | STRAP, MAWD, UNRIP | |||
|
Domain Architecture |

|
|||||
| Description | Serine-threonine kinase receptor-associated protein (UNR-interacting protein) (WD-40 repeat protein PT-WD) (MAP activator with WD repeats). | |||||
|
STRAP_MOUSE
|
||||||
| NC score | 0.005623 (rank : 176) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1Z2 | Gene names | Strap, Unrip | |||
|
Domain Architecture |

|
|||||
| Description | Serine-threonine kinase receptor-associated protein (UNR-interacting protein). | |||||
|
NXF1_HUMAN
|
||||||
| NC score | 0.005372 (rank : 177) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBU9, Q99799, Q9UQL2 | Gene names | NXF1, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear RNA export factor 1 (Tip-associating protein) (Tip-associated protein) (mRNA export factor TAP). | |||||
|
SGOL2_HUMAN
|
||||||
| NC score | 0.005165 (rank : 178) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
NTRI_MOUSE
|
||||||
| NC score | 0.005039 (rank : 179) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PJ0 | Gene names | Nt, Hnt | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrimin precursor. | |||||
|
DUS4_MOUSE
|
||||||
| NC score | 0.004953 (rank : 180) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS4_HUMAN
|
||||||
| NC score | 0.004916 (rank : 181) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
ITA5_HUMAN
|
||||||
| NC score | 0.004873 (rank : 182) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08648, Q96HA5 | Gene names | ITGA5, FNRA | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
CATZ_MOUSE
|
||||||
| NC score | 0.004808 (rank : 183) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUU7 | Gene names | Ctsz | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-). | |||||
|
NTRI_HUMAN
|
||||||
| NC score | 0.004336 (rank : 184) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P121, Q6UXJ3, Q86VJ9 | Gene names | NT | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrimin precursor (hNT). | |||||
|
JHD1B_HUMAN
|
||||||
| NC score | 0.003984 (rank : 185) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHM5, Q8NCI2, Q96HC7, Q96SL0, Q96T03, Q9NS96, Q9UF75 | Gene names | FBXL10, CXXC2, FBL10, JHDM1B, PCCX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10) (Protein JEMMA) (Jumonji domain-containing EMSY-interactor methyltransferase motif protein) (CXXC-type zinc finger protein 2) (Protein-containing CXXC domain 2). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.003552 (rank : 186) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
ADAM9_MOUSE
|
||||||
| NC score | 0.003394 (rank : 187) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61072, Q60618, Q61853, Q80U94 | Gene names | Adam9, Kiaa0021, Mdc9, Mltng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma). | |||||
|
U3IP2_HUMAN
|
||||||
| NC score | 0.003339 (rank : 188) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43818, Q8IZ30 | Gene names | RNU3IP2, U355K | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar RNA-interacting protein 2 (U3 small nucleolar ribonucleoprotein-associated 55 kDa protein) (U3 snoRNP-associated 55 kDa protein) (U3-55K). | |||||
|
PK1IP_MOUSE
|
||||||
| NC score | 0.003228 (rank : 189) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DCE5, Q3UBK7, Q80UT4, Q8C5N6, Q923K2, Q9D3E9 | Gene names | Pak1ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p21-activated protein kinase-interacting protein 1 (PAK1-interacting protein 1) (Putative PAK inhibitor Skb15). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.003185 (rank : 190) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.003082 (rank : 191) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
TAP2_MOUSE
|
||||||
| NC score | 0.002681 (rank : 192) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36371 | Gene names | Tap2, Abcb3, Ham-2, Ham2 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen peptide transporter 2 (APT2) (ATP-binding cassette sub-family B member 3) (Histocompatibility antigen modifier 2). | |||||
|
PLCB4_HUMAN
|
||||||
| NC score | -0.000025 (rank : 193) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
ZNF85_HUMAN
|
||||||
| NC score | -0.003688 (rank : 194) | θ value | 1.06291 (rank : 100) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03923 | Gene names | ZNF85 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 85 (Zinc finger protein HPF4) (HTF1). | |||||
|
PRDM4_MOUSE
|
||||||
| NC score | -0.004521 (rank : 195) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
ZN431_HUMAN
|
||||||
| NC score | -0.004958 (rank : 196) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF32, Q8IWC4 | Gene names | ZNF431, KIAA1969 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 431. | |||||
|
PRDM4_HUMAN
|
||||||
| NC score | -0.005122 (rank : 197) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||