Please be patient as the page loads
|
UBE3A_MOUSE
|
||||||
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBE3A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998778 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | 7.17548e-90 (rank : 3) | NC score | 0.963300 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | 8.77142e-88 (rank : 4) | NC score | 0.961516 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HERC3_HUMAN
|
||||||
| θ value | 7.20884e-74 (rank : 5) | NC score | 0.796720 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HERC5_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 6) | NC score | 0.800945 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 6.56568e-51 (rank : 7) | NC score | 0.871250 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 3.2585e-50 (rank : 8) | NC score | 0.872780 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 6.14528e-49 (rank : 9) | NC score | 0.868769 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 6.14528e-49 (rank : 10) | NC score | 0.869827 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 3.04986e-48 (rank : 11) | NC score | 0.810995 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 3.04986e-48 (rank : 12) | NC score | 0.811993 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 3.04986e-48 (rank : 13) | NC score | 0.825932 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 1.15895e-47 (rank : 14) | NC score | 0.831870 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | 7.51208e-47 (rank : 15) | NC score | 0.910725 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | 9.81109e-47 (rank : 16) | NC score | 0.911028 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 17) | NC score | 0.876996 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 1.67352e-46 (rank : 18) | NC score | 0.869690 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 6.35935e-46 (rank : 19) | NC score | 0.861364 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | 6.35935e-46 (rank : 20) | NC score | 0.868105 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | 2.41655e-45 (rank : 21) | NC score | 0.883291 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | 5.38352e-45 (rank : 22) | NC score | 0.879757 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | 4.55743e-44 (rank : 23) | NC score | 0.878274 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 24) | NC score | 0.831418 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
K0317_HUMAN
|
||||||
| θ value | 1.47289e-26 (rank : 25) | NC score | 0.872461 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 26) | NC score | 0.871725 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
HECD1_HUMAN
|
||||||
| θ value | 2.20094e-22 (rank : 27) | NC score | 0.591843 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 28) | NC score | 0.638624 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| θ value | 9.24701e-21 (rank : 29) | NC score | 0.635475 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 30) | NC score | 0.705426 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 31) | NC score | 0.703672 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
HECD3_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 32) | NC score | 0.733420 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 33) | NC score | 0.728490 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.033497 (rank : 101) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.003486 (rank : 109) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
VOME_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.024173 (rank : 102) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XI7, Q5XK13, Q8CDY1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeromodulin precursor. | |||||
|
CEP55_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.004147 (rank : 107) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.001411 (rank : 111) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | -0.001605 (rank : 113) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
FBX40_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.010399 (rank : 104) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62932 | Gene names | Fbxo40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40. | |||||
|
SEP10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.002665 (rank : 110) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C650, Q7TSA5 | Gene names | Sept10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
SYM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.014106 (rank : 103) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.003902 (rank : 108) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.008039 (rank : 105) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | -0.001317 (rank : 112) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
NARG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.004213 (rank : 106) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXJ9, Q8IWH4, Q8NEV2, Q9H8P6 | Gene names | NARG1, GA19, NATH, TBDN100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Tubedown-1 protein) (Tbdn100) (Gastric cancer antigen Ga19). | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | -0.004791 (rank : 114) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
ALS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.072923 (rank : 76) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.071045 (rank : 78) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
APBB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.087099 (rank : 70) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.083857 (rank : 73) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
APBB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.058831 (rank : 89) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95704, Q9NYX6, Q9NYX7, Q9NYX8 | Gene names | APBB3, FE65L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3 (Fe65-like protein 2) (Fe65L2). | |||||
|
APBB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.059152 (rank : 88) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R1C9 | Gene names | Apbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3. | |||||
|
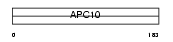
APC10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.050820 (rank : 100) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UM13, Q2V500, Q9UG51, Q9Y5R0 | Gene names | ANAPC10, APC10 | |||
|
Domain Architecture |
|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
BAG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.191537 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.161751 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
CUL7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.063349 (rank : 84) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
DOC2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.051099 (rank : 99) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
DOC2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.053808 (rank : 94) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
FA62A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.061084 (rank : 86) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.072612 (rank : 77) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
GAS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.130432 (rank : 54) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
GAS7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.129932 (rank : 55) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
K1333_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.063358 (rank : 83) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.136538 (rank : 53) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.137051 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.136992 (rank : 52) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.146958 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.058557 (rank : 90) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.056747 (rank : 92) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.208654 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PIN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.207281 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PINL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.248382 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.148040 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.111203 (rank : 61) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.110005 (rank : 64) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RASL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.060070 (rank : 87) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.056734 (rank : 93) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.053330 (rank : 97) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
RCBT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.084541 (rank : 72) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
RCBT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.086814 (rank : 71) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
RCBT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.093012 (rank : 69) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
RCBT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.094067 (rank : 68) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
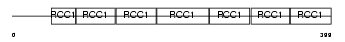
RCC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.110821 (rank : 62) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
RCC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.109389 (rank : 65) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VE37, Q3UDB6 | Gene names | Rcc1, Chc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1). | |||||
|
RCC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.115263 (rank : 58) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
RCC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.114810 (rank : 59) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
RHG12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053503 (rank : 95) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.061468 (rank : 85) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RPGR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.112307 (rank : 60) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
RPGR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.107804 (rank : 67) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
SAV1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.265691 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
SAV1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.266004 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.063568 (rank : 82) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SRGEF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.120229 (rank : 56) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
SRGEF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.116522 (rank : 57) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
STXB4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.080086 (rank : 75) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
STXB4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.080189 (rank : 74) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
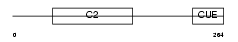
TOLIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.051197 (rank : 98) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H0E2, Q9H9E6, Q9UJ69 | Gene names | TOLLIP | |||
|
Domain Architecture |
|
|||||
| Description | Toll-interacting protein. | |||||
|
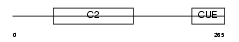
TOLIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.053370 (rank : 96) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZ06, Q543N1, Q9D5P5 | Gene names | Tollip | |||
|
Domain Architecture |
|
|||||
| Description | Toll-interacting protein. | |||||
|
UN13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.065541 (rank : 81) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
UN13B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.068619 (rank : 80) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.068622 (rank : 79) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
WBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.056949 (rank : 91) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
WBS16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.107806 (rank : 66) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96I51, Q548B1, Q9H0G7 | Gene names | WBSCR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein (RCC1-like G exchanging factor-like protein). | |||||
|
WBS16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.110394 (rank : 63) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYF5 | Gene names | Wbscr16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein homolog. | |||||
|
WWC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.222952 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.223490 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
WWOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.206486 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
WWOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.204711 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
WWTR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.177554 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.180222 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.169506 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
YAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.292702 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.998778 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.963300 (rank : 3) | θ value | 7.17548e-90 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.961516 (rank : 4) | θ value | 8.77142e-88 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.911028 (rank : 5) | θ value | 9.81109e-47 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.910725 (rank : 6) | θ value | 7.51208e-47 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.883291 (rank : 7) | θ value | 2.41655e-45 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.879757 (rank : 8) | θ value | 5.38352e-45 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.878274 (rank : 9) | θ value | 4.55743e-44 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.876996 (rank : 10) | θ value | 1.28137e-46 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.872780 (rank : 11) | θ value | 3.2585e-50 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
K0317_HUMAN
|
||||||
| NC score | 0.872461 (rank : 12) | θ value | 1.47289e-26 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| NC score | 0.871725 (rank : 13) | θ value | 4.28545e-26 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.871250 (rank : 14) | θ value | 6.56568e-51 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.869827 (rank : 15) | θ value | 6.14528e-49 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.869690 (rank : 16) | θ value | 1.67352e-46 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.868769 (rank : 17) | θ value | 6.14528e-49 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.868105 (rank : 18) | θ value | 6.35935e-46 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.861364 (rank : 19) | θ value | 6.35935e-46 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.831870 (rank : 20) | θ value | 1.15895e-47 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.831418 (rank : 21) | θ value | 1.52067e-31 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.825932 (rank : 22) | θ value | 3.04986e-48 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.811993 (rank : 23) | θ value | 3.04986e-48 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.810995 (rank : 24) | θ value | 3.04986e-48 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.800945 (rank : 25) | θ value | 8.55514e-59 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.796720 (rank : 26) | θ value | 7.20884e-74 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.733420 (rank : 27) | θ value | 8.38298e-14 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.728490 (rank : 28) | θ value | 5.43371e-13 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.705426 (rank : 29) | θ value | 2.69047e-20 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.703672 (rank : 30) | θ value | 3.51386e-20 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.638624 (rank : 31) | θ value | 4.15078e-21 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.635475 (rank : 32) | θ value | 9.24701e-21 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.591843 (rank : 33) | θ value | 2.20094e-22 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
YAP1_MOUSE
|
||||||
| NC score | 0.292702 (rank : 34) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
SAV1_MOUSE
|
||||||
| NC score | 0.266004 (rank : 35) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
SAV1_HUMAN
|
||||||
| NC score | 0.265691 (rank : 36) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
PINL_HUMAN
|
||||||
| NC score | 0.248382 (rank : 37) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.223490 (rank : 38) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
WWC1_HUMAN
|
||||||
| NC score | 0.222952 (rank : 39) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
PIN1_HUMAN
|
||||||
| NC score | 0.208654 (rank : 40) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PIN1_MOUSE
|
||||||
| NC score | 0.207281 (rank : 41) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
WWOX_HUMAN
|
||||||
| NC score | 0.206486 (rank : 42) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
WWOX_MOUSE
|
||||||
| NC score | 0.204711 (rank : 43) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.191537 (rank : 44) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
WWTR1_MOUSE
|
||||||
| NC score | 0.180222 (rank : 45) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_HUMAN
|
||||||
| NC score | 0.177554 (rank : 46) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.169506 (rank : 47) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.161751 (rank : 48) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.148040 (rank : 49) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.146958 (rank : 50) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.137051 (rank : 51) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.136992 (rank : 52) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.136538 (rank : 53) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
GAS7_HUMAN
|
||||||
| NC score | 0.130432 (rank : 54) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
GAS7_MOUSE
|
||||||
| NC score | 0.129932 (rank : 55) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
SRGEF_HUMAN
|
||||||
| NC score | 0.120229 (rank : 56) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
SRGEF_MOUSE
|
||||||
| NC score | 0.116522 (rank : 57) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
RCC2_HUMAN
|
||||||
| NC score | 0.115263 (rank : 58) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
RCC2_MOUSE
|
||||||
| NC score | 0.114810 (rank : 59) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
RPGR_HUMAN
|
||||||
| NC score | 0.112307 (rank : 60) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.111203 (rank : 61) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
RCC1_HUMAN
|
||||||
| NC score | 0.110821 (rank : 62) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
WBS16_MOUSE
|
||||||
| NC score | 0.110394 (rank : 63) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYF5 | Gene names | Wbscr16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein homolog. | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.110005 (rank : 64) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RCC1_MOUSE
|
||||||
| NC score | 0.109389 (rank : 65) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VE37, Q3UDB6 | Gene names | Rcc1, Chc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1). | |||||
|
WBS16_HUMAN
|
||||||
| NC score | 0.107806 (rank : 66) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96I51, Q548B1, Q9H0G7 | Gene names | WBSCR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein (RCC1-like G exchanging factor-like protein). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.107804 (rank : 67) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
RCBT2_MOUSE
|
||||||
| NC score | 0.094067 (rank : 68) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
RCBT2_HUMAN
|
||||||
| NC score | 0.093012 (rank : 69) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.087099 (rank : 70) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
RCBT1_MOUSE
|
||||||
| NC score | 0.086814 (rank : 71) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
RCBT1_HUMAN
|
||||||
| NC score | 0.084541 (rank : 72) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.083857 (rank : 73) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
STXB4_MOUSE
|
||||||
| NC score | 0.080189 (rank : 74) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
STXB4_HUMAN
|
||||||
| NC score | 0.080086 (rank : 75) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.072923 (rank : 76) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.072612 (rank : 77) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.071045 (rank : 78) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
UN13B_MOUSE
|
||||||
| NC score | 0.068622 (rank : 79) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13B_HUMAN
|
||||||
| NC score | 0.068619 (rank : 80) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.065541 (rank : 81) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.063568 (rank : 82) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
K1333_MOUSE
|
||||||
| NC score | 0.063358 (rank : 83) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.063349 (rank : 84) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.061468 (rank : 85) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.061084 (rank : 86) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
RASL1_HUMAN
|
||||||
| NC score | 0.060070 (rank : 87) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
APBB3_MOUSE
|
||||||
| NC score | 0.059152 (rank : 88) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R1C9 | Gene names | Apbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3. | |||||
|
APBB3_HUMAN
|
||||||
| NC score | 0.058831 (rank : 89) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95704, Q9NYX6, Q9NYX7, Q9NYX8 | Gene names | APBB3, FE65L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3 (Fe65-like protein 2) (Fe65L2). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.058557 (rank : 90) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
WBP4_MOUSE
|
||||||
| NC score | 0.056949 (rank : 91) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.056747 (rank : 92) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
RASL1_MOUSE
|
||||||
| NC score | 0.056734 (rank : 93) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
DOC2A_MOUSE
|
||||||
| NC score | 0.053808 (rank : 94) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.053503 (rank : 95) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
TOLIP_MOUSE
|
||||||
| NC score | 0.053370 (rank : 96) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZ06, Q543N1, Q9D5P5 | Gene names | Tollip | |||
|
Domain Architecture |

|
|||||
| Description | Toll-interacting protein. | |||||
|
RASL2_HUMAN
|
||||||
| NC score | 0.053330 (rank : 97) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
TOLIP_HUMAN
|
||||||
| NC score | 0.051197 (rank : 98) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H0E2, Q9H9E6, Q9UJ69 | Gene names | TOLLIP | |||
|
Domain Architecture |

|
|||||
| Description | Toll-interacting protein. | |||||
|
DOC2A_HUMAN
|
||||||
| NC score | 0.051099 (rank : 99) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
APC10_HUMAN
|
||||||
| NC score | 0.050820 (rank : 100) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UM13, Q2V500, Q9UG51, Q9Y5R0 | Gene names | ANAPC10, APC10 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.033497 (rank : 101) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
VOME_MOUSE
|
||||||
| NC score | 0.024173 (rank : 102) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XI7, Q5XK13, Q8CDY1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeromodulin precursor. | |||||
|
SYM_MOUSE
|
||||||
| NC score | 0.014106 (rank : 103) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
FBX40_MOUSE
|
||||||
| NC score | 0.010399 (rank : 104) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62932 | Gene names | Fbxo40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.008039 (rank : 105) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
NARG1_HUMAN
|
||||||
| NC score | 0.004213 (rank : 106) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXJ9, Q8IWH4, Q8NEV2, Q9H8P6 | Gene names | NARG1, GA19, NATH, TBDN100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Tubedown-1 protein) (Tbdn100) (Gastric cancer antigen Ga19). | |||||
|
CEP55_HUMAN
|
||||||
| NC score | 0.004147 (rank : 107) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.003902 (rank : 108) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.003486 (rank : 109) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SEP10_MOUSE
|
||||||
| NC score | 0.002665 (rank : 110) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C650, Q7TSA5 | Gene names | Sept10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.001411 (rank : 111) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
CP250_MOUSE
|
||||||
| NC score | -0.001317 (rank : 112) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CP250_HUMAN
|
||||||
| NC score | -0.001605 (rank : 113) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | -0.004791 (rank : 114) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||