Please be patient as the page loads
|
APBB2_MOUSE
|
||||||
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
APBB2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987777 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
APBB1_MOUSE
|
||||||
| θ value | 1.19589e-169 (rank : 3) | NC score | 0.954673 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
APBB1_HUMAN
|
||||||
| θ value | 5.02443e-168 (rank : 4) | NC score | 0.956712 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
APBB3_HUMAN
|
||||||
| θ value | 1.48926e-103 (rank : 5) | NC score | 0.940212 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95704, Q9NYX6, Q9NYX7, Q9NYX8 | Gene names | APBB3, FE65L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3 (Fe65-like protein 2) (Fe65L2). | |||||
|
APBB3_MOUSE
|
||||||
| θ value | 2.80862e-102 (rank : 6) | NC score | 0.940102 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R1C9 | Gene names | Apbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3. | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 7) | NC score | 0.182589 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 8) | NC score | 0.169774 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.150578 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 10) | NC score | 0.049215 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 11) | NC score | 0.150765 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 12) | NC score | 0.147991 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.115746 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 14) | NC score | 0.146833 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 15) | NC score | 0.145549 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 16) | NC score | 0.161005 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.166925 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 18) | NC score | 0.143461 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.046852 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.032992 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ARH_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.122806 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SW96, Q6TQS9, Q8N2Y0, Q9UFI9 | Gene names | LDLRAP1, ARH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.141049 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.147451 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
ARH_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.122247 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C142, Q6NWV6, Q8VDQ0 | Gene names | Ldlrap1, Arh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein homolog). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.099885 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.030458 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
GAS7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.116324 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
CAPON_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.112257 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75052, O43564 | Gene names | NOS1AP, CAPON, KIAA0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.081252 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
NUMBL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.090044 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.021122 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.035345 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.026175 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.045659 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.010633 (rank : 104) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
CAPON_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.103163 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D3A8, Q80TZ6 | Gene names | Nos1ap, Capon, Kiaa0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
PIN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.158335 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PIN1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.176804 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.118863 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
GAS7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.103266 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.053020 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
KLH14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.008111 (rank : 106) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.040855 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ZEP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.007075 (rank : 111) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.032427 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PINL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.179114 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
CA106_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.020347 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
CI079_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.053841 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.005172 (rank : 116) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
NUMB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.097242 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
NUMB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.098448 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
STON2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.031587 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.007169 (rank : 110) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.023791 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.007854 (rank : 107) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.029833 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZN451_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.010142 (rank : 105) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C0P7 | Gene names | Znf451, Zfp451 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.028640 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.018468 (rank : 94) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CD44_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.023743 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16070, P22511, Q04858, Q13419, Q13957, Q13958, Q13959, Q13960, Q13961, Q13967, Q13968, Q13980, Q15861, Q16064, Q16065, Q16066, Q16208, Q16522, Q96J24 | Gene names | CD44, LHR | |||
|
Domain Architecture |
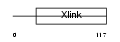
|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein I) (PGP-1) (HUTCH-I) (Extracellular matrix receptor-III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Heparan sulfate proteoglycan) (Epican) (CDw44). | |||||
|
GP125_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.004844 (rank : 117) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT36, Q6PE67, Q8VE71 | Gene names | Gpr125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
IQGA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.011148 (rank : 102) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.018811 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
THYG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.011557 (rank : 101) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.027217 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.023929 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.001704 (rank : 119) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.016898 (rank : 96) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
GP1BA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.013229 (rank : 98) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
MYOM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.005847 (rank : 114) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62234 | Gene names | Myom1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (Skelemin). | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.007815 (rank : 108) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
ZN507_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.003073 (rank : 118) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TCN5 | Gene names | ZNF507 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 507. | |||||
|
CASZ1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.011759 (rank : 99) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
CSTF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.005714 (rank : 115) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |
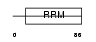
|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.023587 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.017763 (rank : 95) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.024822 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
EME1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.011108 (rank : 103) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJW7 | Gene names | Eme1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crossover junction endonuclease EME1 (EC 3.1.22.-). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.018569 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
GNB1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.013421 (rank : 97) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYB4, Q9H2S2, Q9H4M4 | Gene names | GNB1L, GY2, KIAA1645, WDR14 | |||
|
Domain Architecture |
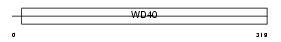
|
|||||
| Description | Guanine nucleotide-binding protein subunit beta-like protein 1 (G protein subunit beta-like protein 1) (WD40 repeat-containing protein deleted in VCFS) (Protein WDVCF) (DGCRK3). | |||||
|
JPH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.006752 (rank : 112) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.000970 (rank : 121) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.022693 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.007450 (rank : 109) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
SOX9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.011745 (rank : 100) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.020323 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.006675 (rank : 113) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.001139 (rank : 120) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
BAG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.056511 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.059588 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.059537 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.055357 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.087164 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.087814 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.060094 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.061147 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.056604 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.057424 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.070636 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HERC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.072524 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.090822 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.099533 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
K0317_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.091944 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.091876 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.056048 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.054686 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
SAV1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.081765 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
SAV1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.077971 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.107067 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.079614 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.084922 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.083857 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.083728 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.083440 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
WWC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.056252 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.055853 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
WWOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.058074 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
WWOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.057548 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
WWTR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.059350 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.082890 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
YAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.095418 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.987777 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
APBB1_HUMAN
|
||||||
| NC score | 0.956712 (rank : 3) | θ value | 5.02443e-168 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
APBB1_MOUSE
|
||||||
| NC score | 0.954673 (rank : 4) | θ value | 1.19589e-169 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
APBB3_HUMAN
|
||||||
| NC score | 0.940212 (rank : 5) | θ value | 1.48926e-103 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95704, Q9NYX6, Q9NYX7, Q9NYX8 | Gene names | APBB3, FE65L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3 (Fe65-like protein 2) (Fe65L2). | |||||
|
APBB3_MOUSE
|
||||||
| NC score | 0.940102 (rank : 6) | θ value | 2.80862e-102 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R1C9 | Gene names | Apbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3. | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.182589 (rank : 7) | θ value | 4.1701e-05 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
PINL_HUMAN
|
||||||
| NC score | 0.179114 (rank : 8) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
PIN1_MOUSE
|
||||||
| NC score | 0.176804 (rank : 9) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.169774 (rank : 10) | θ value | 0.00035302 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.166925 (rank : 11) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.161005 (rank : 12) | θ value | 0.0113563 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
PIN1_HUMAN
|
||||||
| NC score | 0.158335 (rank : 13) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.150765 (rank : 14) | θ value | 0.00509761 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.150578 (rank : 15) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.147991 (rank : 16) | θ value | 0.00665767 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.147451 (rank : 17) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.146833 (rank : 18) | θ value | 0.0113563 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.145549 (rank : 19) | θ value | 0.0113563 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.143461 (rank : 20) | θ value | 0.0330416 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.141049 (rank : 21) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
ARH_HUMAN
|
||||||
| NC score | 0.122806 (rank : 22) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SW96, Q6TQS9, Q8N2Y0, Q9UFI9 | Gene names | LDLRAP1, ARH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein). | |||||
|
ARH_MOUSE
|
||||||
| NC score | 0.122247 (rank : 23) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C142, Q6NWV6, Q8VDQ0 | Gene names | Ldlrap1, Arh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein homolog). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.118863 (rank : 24) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
GAS7_HUMAN
|
||||||
| NC score | 0.116324 (rank : 25) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.115746 (rank : 26) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
CAPON_HUMAN
|
||||||
| NC score | 0.112257 (rank : 27) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75052, O43564 | Gene names | NOS1AP, CAPON, KIAA0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.107067 (rank : 28) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
GAS7_MOUSE
|
||||||
| NC score | 0.103266 (rank : 29) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
CAPON_MOUSE
|
||||||
| NC score | 0.103163 (rank : 30) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D3A8, Q80TZ6 | Gene names | Nos1ap, Capon, Kiaa0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.099885 (rank : 31) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.099533 (rank : 32) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
NUMB_MOUSE
|
||||||
| NC score | 0.098448 (rank : 33) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.097242 (rank : 34) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
YAP1_MOUSE
|
||||||
| NC score | 0.095418 (rank : 35) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
K0317_HUMAN
|
||||||
| NC score | 0.091944 (rank : 36) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| NC score | 0.091876 (rank : 37) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.090822 (rank : 38) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
NUMBL_MOUSE
|
||||||
| NC score | 0.090044 (rank : 39) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.087814 (rank : 40) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.087164 (rank : 41) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.084922 (rank : 42) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.083857 (rank : 43) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.083728 (rank : 44) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.083440 (rank : 45) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.082890 (rank : 46) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
SAV1_HUMAN
|
||||||
| NC score | 0.081765 (rank : 47) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.081252 (rank : 48) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.079614 (rank : 49) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
SAV1_MOUSE
|
||||||
| NC score | 0.077971 (rank : 50) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.072524 (rank : 51) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.070636 (rank : 52) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.061147 (rank : 53) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.060094 (rank : 54) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.059588 (rank : 55) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.059537 (rank : 56) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
WWTR1_HUMAN
|
||||||
| NC score | 0.059350 (rank : 57) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWOX_HUMAN
|
||||||
| NC score | 0.058074 (rank : 58) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
WWOX_MOUSE
|
||||||
| NC score | 0.057548 (rank : 59) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.057424 (rank : 60) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.056604 (rank : 61) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.056511 (rank : 62) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
WWC1_HUMAN
|
||||||
| NC score | 0.056252 (rank : 63) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.056048 (rank : 64) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.055853 (rank : 65) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.055357 (rank : 66) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.054686 (rank : 67) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.053841 (rank : 68) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.053020 (rank : 69) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.049215 (rank : 70) | θ value | 0.00228821 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.046852 (rank : 71) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.045659 (rank : 72) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.040855 (rank : 73) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.035345 (rank : 74) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.032992 (rank : 75) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.032427 (rank : 76) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
STON2_MOUSE
|
||||||
| NC score | 0.031587 (rank : 77) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.030458 (rank : 78) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.029833 (rank : 79) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.028640 (rank : 80) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.027217 (rank : 81) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.026175 (rank : 82) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.024822 (rank : 83) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.023929 (rank : 84) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.023791 (rank : 85) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
CD44_HUMAN
|
||||||
| NC score | 0.023743 (rank : 86) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16070, P22511, Q04858, Q13419, Q13957, Q13958, Q13959, Q13960, Q13961, Q13967, Q13968, Q13980, Q15861, Q16064, Q16065, Q16066, Q16208, Q16522, Q96J24 | Gene names | CD44, LHR | |||
|
Domain Architecture |

|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein I) (PGP-1) (HUTCH-I) (Extracellular matrix receptor-III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Heparan sulfate proteoglycan) (Epican) (CDw44). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.023587 (rank : 87) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.022693 (rank : 88) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.021122 (rank : 89) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
CA106_MOUSE
|
||||||
| NC score | 0.020347 (rank : 90) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.020323 (rank : 91) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.018811 (rank : 92) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.018569 (rank : 93) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.018468 (rank : 94) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.017763 (rank : 95) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.016898 (rank : 96) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
GNB1L_HUMAN
|
||||||
| NC score | 0.013421 (rank : 97) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYB4, Q9H2S2, Q9H4M4 | Gene names | GNB1L, GY2, KIAA1645, WDR14 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein subunit beta-like protein 1 (G protein subunit beta-like protein 1) (WD40 repeat-containing protein deleted in VCFS) (Protein WDVCF) (DGCRK3). | |||||
|
GP1BA_HUMAN
|
||||||
| NC score | 0.013229 (rank : 98) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
CASZ1_HUMAN
|
||||||
| NC score | 0.011759 (rank : 99) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
SOX9_MOUSE
|
||||||
| NC score | 0.011745 (rank : 100) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.011557 (rank : 101) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
IQGA3_HUMAN
|
||||||
| NC score | 0.011148 (rank : 102) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||
|
EME1_MOUSE
|
||||||
| NC score | 0.011108 (rank : 103) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJW7 | Gene names | Eme1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crossover junction endonuclease EME1 (EC 3.1.22.-). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.010633 (rank : 104) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
ZN451_MOUSE
|
||||||
| NC score | 0.010142 (rank : 105) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C0P7 | Gene names | Znf451, Zfp451 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451. | |||||
|
KLH14_HUMAN
|
||||||
| NC score | 0.008111 (rank : 106) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.007854 (rank : 107) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.007815 (rank : 108) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.007450 (rank : 109) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.007169 (rank : 110) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
ZEP2_HUMAN
|
||||||
| NC score | 0.007075 (rank : 111) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
JPH1_MOUSE
|
||||||
| NC score | 0.006752 (rank : 112) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.006675 (rank : 113) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
MYOM1_MOUSE
|
||||||
| NC score | 0.005847 (rank : 114) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62234 | Gene names | Myom1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (Skelemin). | |||||
|
CSTF2_HUMAN
|
||||||
| NC score | 0.005714 (rank : 115) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.005172 (rank : 116) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
GP125_MOUSE
|
||||||
| NC score | 0.004844 (rank : 117) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT36, Q6PE67, Q8VE71 | Gene names | Gpr125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
ZN507_HUMAN
|
||||||
| NC score | 0.003073 (rank : 118) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TCN5 | Gene names | ZNF507 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 507. | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.001704 (rank : 119) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.001139 (rank : 120) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.000970 (rank : 121) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||