Please be patient as the page loads
|
SAV1_MOUSE
|
||||||
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SAV1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998967 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
SAV1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 3) | NC score | 0.461672 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 4) | NC score | 0.469224 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 5) | NC score | 0.436515 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 6) | NC score | 0.437451 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 7) | NC score | 0.525672 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 8) | NC score | 0.528728 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 9) | NC score | 0.528156 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 10) | NC score | 0.533664 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 11) | NC score | 0.530832 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 12) | NC score | 0.531921 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
WWC1_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 13) | NC score | 0.542652 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 14) | NC score | 0.540240 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 15) | NC score | 0.477033 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 16) | NC score | 0.469811 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 17) | NC score | 0.470361 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
WWOX_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 18) | NC score | 0.449809 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
WWOX_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 19) | NC score | 0.445957 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 20) | NC score | 0.473214 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
YAP1_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 21) | NC score | 0.603615 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 22) | NC score | 0.339723 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 23) | NC score | 0.473772 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 24) | NC score | 0.473292 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 25) | NC score | 0.454782 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
GAS7_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 26) | NC score | 0.299722 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
WWTR1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 27) | NC score | 0.462862 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 28) | NC score | 0.466323 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
GAS7_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 29) | NC score | 0.286773 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 30) | NC score | 0.446855 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
STXB4_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 31) | NC score | 0.234997 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
STXB4_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 32) | NC score | 0.236583 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
BAG3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 33) | NC score | 0.362565 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 34) | NC score | 0.317286 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
PIN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.336782 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PIN1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.340021 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PINL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.387810 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.005462 (rank : 120) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
APBB3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.086904 (rank : 63) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95704, Q9NYX6, Q9NYX7, Q9NYX8 | Gene names | APBB3, FE65L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3 (Fe65-like protein 2) (Fe65L2). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.002166 (rank : 122) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
STK4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.002854 (rank : 121) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13043, Q15802, Q4G156, Q5H982, Q6PD60, Q9BR32, Q9NTZ4 | Gene names | STK4, MST1 | |||
|
Domain Architecture |
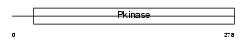
|
|||||
| Description | Serine/threonine-protein kinase 4 (EC 2.7.11.1) (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1) (Serine/threonine-protein kinase Krs-2). | |||||
|
TRIP6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.012216 (rank : 119) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
STK3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.002056 (rank : 124) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 869 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JI10, Q60877, Q80UG4, Q8CI58 | Gene names | Stk3, Mess1, Mst2 | |||
|
Domain Architecture |
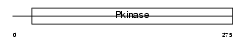
|
|||||
| Description | Serine/threonine-protein kinase 3 (EC 2.7.11.1) (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2). | |||||
|
STK4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.002095 (rank : 123) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JI11 | Gene names | Stk4, Mst1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 4 (EC 2.7.11.1) (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.018473 (rank : 118) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.036383 (rank : 115) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.033623 (rank : 116) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.029799 (rank : 117) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
APBA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.050472 (rank : 111) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |
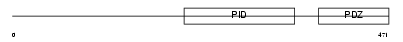
|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
APBB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.094256 (rank : 61) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.077971 (rank : 64) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
APBB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.071540 (rank : 65) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R1C9 | Gene names | Apbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3. | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.060427 (rank : 81) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.060549 (rank : 80) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
DHRSX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.053837 (rank : 96) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N5I4, Q6UWC7, Q8WUS4, Q96GR8, Q9NTF6 | Gene names | DHRSX, CXorf11, DHRS5X | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member on chromosome X precursor (EC 1.1.-.-) (DHRSXY). | |||||
|
DRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.061809 (rank : 74) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
EDD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.190589 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.190821 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.052256 (rank : 103) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
GRIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.055582 (rank : 92) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GRIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.052806 (rank : 101) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.056342 (rank : 91) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.173769 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.276688 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.278289 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.191355 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.194712 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.179896 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.181978 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.225043 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HERC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.230934 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.281502 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.281955 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
IL16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.053313 (rank : 97) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
INADL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.060175 (rank : 82) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
INADL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.060584 (rank : 79) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
K0317_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.291260 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.291089 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K1849_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.070382 (rank : 66) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
K1849_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.063964 (rank : 72) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
LIN7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.057446 (rank : 88) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
LIN7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.057416 (rank : 89) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
LIN7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.059564 (rank : 83) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
LIN7B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.060692 (rank : 78) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
LIN7C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.061127 (rank : 76) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.061127 (rank : 77) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LNX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.052405 (rank : 102) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N448, Q5W0P0, Q6ZMH2, Q96SH4 | Gene names | LNX2, PDZRN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2) (PDZ domain- containing RING finger protein 1). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.065491 (rank : 70) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.061179 (rank : 75) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
NHERF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.050260 (rank : 112) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |
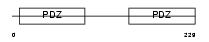
|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.052838 (rank : 100) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.059263 (rank : 84) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
NHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.059090 (rank : 85) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
PAR3L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.058569 (rank : 86) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
PARD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.065693 (rank : 69) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
PARD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.066797 (rank : 68) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
PDLI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.052962 (rank : 98) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JY6, Q7Z584, Q86WM8, Q8WZ29, Q9H4L9, Q9H7I2 | Gene names | PDLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique) (PDZ-LIM protein). | |||||
|
PDZ11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.051182 (rank : 108) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.050089 (rank : 114) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.054490 (rank : 93) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.057157 (rank : 90) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.054322 (rank : 94) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
PRP40_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.115755 (rank : 58) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.112504 (rank : 59) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
PSCBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050654 (rank : 110) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60759, Q15630, Q8NE32 | Gene names | PSCDBP, CYTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (CYBR) (Cytohesin binder and regulator) (Cytohesin-interacting protein). | |||||
|
RDH12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.050171 (rank : 113) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NR8, Q8TAW6 | Gene names | RDH12 | |||
|
Domain Architecture |
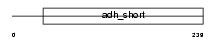
|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-) (All-trans and 9-cis retinol dehydrogenase). | |||||
|
RDH12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.051781 (rank : 106) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |
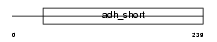
|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
RDH14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.051895 (rank : 104) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBH5 | Gene names | RDH14, PAN2 | |||
|
Domain Architecture |
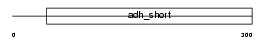
|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
RDH14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.051308 (rank : 107) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERI6 | Gene names | Rdh14 | |||
|
Domain Architecture |
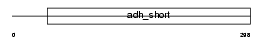
|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
RHG12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.058126 (rank : 87) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.068989 (rank : 67) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
SETD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.093543 (rank : 62) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SYJ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.065357 (rank : 71) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.250125 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.269552 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.266004 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.266006 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.265007 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
WBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.062968 (rank : 73) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051060 (rank : 109) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.052872 (rank : 99) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
WWC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.105043 (rank : 60) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULE0, Q659C1, Q9BTQ1 | Gene names | WWC3, KIAA1280 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WWC family member 3. | |||||
|
ZO3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.053881 (rank : 95) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ZO3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.051875 (rank : 105) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
SAV1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
SAV1_HUMAN
|
||||||
| NC score | 0.998967 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
YAP1_MOUSE
|
||||||
| NC score | 0.603615 (rank : 3) | θ value | 1.53129e-07 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
WWC1_HUMAN
|
||||||
| NC score | 0.542652 (rank : 4) | θ value | 2.13673e-09 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.540240 (rank : 5) | θ value | 2.13673e-09 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.533664 (rank : 6) | θ value | 1.74796e-11 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.531921 (rank : 7) | θ value | 5.08577e-11 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.530832 (rank : 8) | θ value | 5.08577e-11 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.528728 (rank : 9) | θ value | 7.84624e-12 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.528156 (rank : 10) | θ value | 1.74796e-11 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.525672 (rank : 11) | θ value | 7.84624e-12 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.477033 (rank : 12) | θ value | 4.76016e-09 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.473772 (rank : 13) | θ value | 1.69304e-06 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.473292 (rank : 14) | θ value | 7.1131e-05 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.473214 (rank : 15) | θ value | 6.87365e-08 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.470361 (rank : 16) | θ value | 8.11959e-09 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.469811 (rank : 17) | θ value | 8.11959e-09 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.469224 (rank : 18) | θ value | 1.09485e-13 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
WWTR1_MOUSE
|
||||||
| NC score | 0.466323 (rank : 19) | θ value | 0.000786445 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_HUMAN
|
||||||
| NC score | 0.462862 (rank : 20) | θ value | 0.000602161 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.461672 (rank : 21) | θ value | 1.09485e-13 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.454782 (rank : 22) | θ value | 0.00020696 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
WWOX_HUMAN
|
||||||
| NC score | 0.449809 (rank : 23) | θ value | 3.08544e-08 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.446855 (rank : 24) | θ value | 0.00509761 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
WWOX_MOUSE
|
||||||
| NC score | 0.445957 (rank : 25) | θ value | 3.08544e-08 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.437451 (rank : 26) | θ value | 3.52202e-12 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.436515 (rank : 27) | θ value | 2.69671e-12 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
PINL_HUMAN
|
||||||
| NC score | 0.387810 (rank : 28) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.362565 (rank : 29) | θ value | 0.0148317 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
PIN1_MOUSE
|
||||||
| NC score | 0.340021 (rank : 30) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.339723 (rank : 31) | θ value | 1.69304e-06 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PIN1_HUMAN
|
||||||
| NC score | 0.336782 (rank : 32) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.317286 (rank : 33) | θ value | 0.0148317 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
GAS7_HUMAN
|
||||||
| NC score | 0.299722 (rank : 34) | θ value | 0.000270298 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
K0317_HUMAN
|
||||||
| NC score | 0.291260 (rank : 35) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| NC score | 0.291089 (rank : 36) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
GAS7_MOUSE
|
||||||
| NC score | 0.286773 (rank : 37) | θ value | 0.00390308 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.281955 (rank : 38) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.281502 (rank : 39) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.278289 (rank : 40) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.276688 (rank : 41) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.269552 (rank : 42) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.266006 (rank : 43) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.266004 (rank : 44) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.265007 (rank : 45) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.250125 (rank : 46) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
STXB4_MOUSE
|
||||||
| NC score | 0.236583 (rank : 47) | θ value | 0.00665767 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
STXB4_HUMAN
|
||||||
| NC score | 0.234997 (rank : 48) | θ value | 0.00665767 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.230934 (rank : 49) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.225043 (rank : 50) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.194712 (rank : 51) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.191355 (rank : 52) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.190821 (rank : 53) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.190589 (rank : 54) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.181978 (rank : 55) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.179896 (rank : 56) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.173769 (rank : 57) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.115755 (rank : 58) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.112504 (rank : 59) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
WWC3_HUMAN
|
||||||
| NC score | 0.105043 (rank : 60) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULE0, Q659C1, Q9BTQ1 | Gene names | WWC3, KIAA1280 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WWC family member 3. | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.094256 (rank : 61) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.093543 (rank : 62) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
APBB3_HUMAN
|
||||||
| NC score | 0.086904 (rank : 63) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95704, Q9NYX6, Q9NYX7, Q9NYX8 | Gene names | APBB3, FE65L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3 (Fe65-like protein 2) (Fe65L2). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.077971 (rank : 64) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
APBB3_MOUSE
|
||||||
| NC score | 0.071540 (rank : 65) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R1C9 | Gene names | Apbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3. | |||||
|
K1849_HUMAN
|
||||||
| NC score | 0.070382 (rank : 66) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.068989 (rank : 67) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
PARD3_MOUSE
|
||||||
| NC score | 0.066797 (rank : 68) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
PARD3_HUMAN
|
||||||
| NC score | 0.065693 (rank : 69) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.065491 (rank : 70) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
SYJ2B_HUMAN
|
||||||
| NC score | 0.065357 (rank : 71) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
K1849_MOUSE
|
||||||
| NC score | 0.063964 (rank : 72) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
WBP4_MOUSE
|
||||||
| NC score | 0.062968 (rank : 73) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
DRP2_HUMAN
|
||||||
| NC score | 0.061809 (rank : 74) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.061179 (rank : 75) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
LIN7C_HUMAN
|
||||||
| NC score | 0.061127 (rank : 76) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| NC score | 0.061127 (rank : 77) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7B_MOUSE
|
||||||
| NC score | 0.060692 (rank : 78) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.060584 (rank : 79) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.060549 (rank : 80) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.060427 (rank : 81) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.060175 (rank : 82) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
LIN7B_HUMAN
|
||||||
| NC score | 0.059564 (rank : 83) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
NHRF2_HUMAN
|
||||||
| NC score | 0.059263 (rank : 84) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
NHRF2_MOUSE
|
||||||
| NC score | 0.059090 (rank : 85) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
PAR3L_HUMAN
|
||||||
| NC score | 0.058569 (rank : 86) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.058126 (rank : 87) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
LIN7A_HUMAN
|
||||||
| NC score | 0.057446 (rank : 88) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
LIN7A_MOUSE
|
||||||
| NC score | 0.057416 (rank : 89) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
PDZD1_MOUSE
|
||||||
| NC score | 0.057157 (rank : 90) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.056342 (rank : 91) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
GRIP1_HUMAN
|
||||||
| NC score | 0.055582 (rank : 92) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.054490 (rank : 93) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZD7_HUMAN
|
||||||
| NC score | 0.054322 (rank : 94) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
ZO3_HUMAN
|
||||||
| NC score | 0.053881 (rank : 95) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
DHRSX_HUMAN
|
||||||
| NC score | 0.053837 (rank : 96) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N5I4, Q6UWC7, Q8WUS4, Q96GR8, Q9NTF6 | Gene names | DHRSX, CXorf11, DHRS5X | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member on chromosome X precursor (EC 1.1.-.-) (DHRSXY). | |||||
|
IL16_HUMAN
|
||||||
| NC score | 0.053313 (rank : 97) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
PDLI2_HUMAN
|
||||||
| NC score | 0.052962 (rank : 98) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JY6, Q7Z584, Q86WM8, Q8WZ29, Q9H4L9, Q9H7I2 | Gene names | PDLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique) (PDZ-LIM protein). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.052872 (rank : 99) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.052838 (rank : 100) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
GRIP1_MOUSE
|
||||||
| NC score | 0.052806 (rank : 101) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
LNX2_HUMAN
|
||||||
| NC score | 0.052405 (rank : 102) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N448, Q5W0P0, Q6ZMH2, Q96SH4 | Gene names | LNX2, PDZRN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2) (PDZ domain- containing RING finger protein 1). | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.052256 (rank : 103) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
RDH14_HUMAN
|
||||||
| NC score | 0.051895 (rank : 104) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBH5 | Gene names | RDH14, PAN2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
ZO3_MOUSE
|
||||||
| NC score | 0.051875 (rank : 105) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
RDH12_MOUSE
|
||||||
| NC score | 0.051781 (rank : 106) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
RDH14_MOUSE
|
||||||
| NC score | 0.051308 (rank : 107) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERI6 | Gene names | Rdh14 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
PDZ11_HUMAN
|
||||||
| NC score | 0.051182 (rank : 108) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.051060 (rank : 109) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
PSCBP_HUMAN
|
||||||
| NC score | 0.050654 (rank : 110) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60759, Q15630, Q8NE32 | Gene names | PSCDBP, CYTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (CYBR) (Cytohesin binder and regulator) (Cytohesin-interacting protein). | |||||
|
APBA3_MOUSE
|
||||||
| NC score | 0.050472 (rank : 111) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
NHERF_HUMAN
|
||||||
| NC score | 0.050260 (rank : 112) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
RDH12_HUMAN
|
||||||
| NC score | 0.050171 (rank : 113) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NR8, Q8TAW6 | Gene names | RDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-) (All-trans and 9-cis retinol dehydrogenase). | |||||
|
PDZ11_MOUSE
|
||||||
| NC score | 0.050089 (rank : 114) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.036383 (rank : 115) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.033623 (rank : 116) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.029799 (rank : 117) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.018473 (rank : 118) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
TRIP6_HUMAN
|
||||||
| NC score | 0.012216 (rank : 119) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.005462 (rank : 120) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
STK4_HUMAN
|
||||||
| NC score | 0.002854 (rank : 121) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13043, Q15802, Q4G156, Q5H982, Q6PD60, Q9BR32, Q9NTZ4 | Gene names | STK4, MST1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 4 (EC 2.7.11.1) (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1) (Serine/threonine-protein kinase Krs-2). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.002166 (rank : 122) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
STK4_MOUSE
|
||||||
| NC score | 0.002095 (rank : 123) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JI11 | Gene names | Stk4, Mst1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 4 (EC 2.7.11.1) (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1). | |||||
|
STK3_MOUSE
|
||||||
| NC score | 0.002056 (rank : 124) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 869 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JI10, Q60877, Q80UG4, Q8CI58 | Gene names | Stk3, Mess1, Mst2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 3 (EC 2.7.11.1) (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2). | |||||