Please be patient as the page loads
|
NARG1_HUMAN
|
||||||
| SwissProt Accessions | Q9BXJ9, Q8IWH4, Q8NEV2, Q9H8P6 | Gene names | NARG1, GA19, NATH, TBDN100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Tubedown-1 protein) (Tbdn100) (Gastric cancer antigen Ga19). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NARG1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q9BXJ9, Q8IWH4, Q8NEV2, Q9H8P6 | Gene names | NARG1, GA19, NATH, TBDN100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Tubedown-1 protein) (Tbdn100) (Gastric cancer antigen Ga19). | |||||
|
NARG1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998997 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q80UM3, Q811Z9, Q9JID5 | Gene names | Narg1, Nat1, Tbdn-1, Tubedown | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase 1) (Protein Tubedown-1). | |||||
|
NARGL_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.959732 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
NARGL_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.969679 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
IFT88_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.162828 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
IFT88_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.171551 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
CRNL1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.127841 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63154, Q542E8, Q9CQC1 | Gene names | Crnkl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crooked neck-like protein 1 (Crooked neck homolog). | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.158588 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.043014 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
BBS4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.165661 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.079123 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
BBS4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.166736 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.040465 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
CRNL1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.122229 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZJ0, Q5JY64, Q8WYI5, Q9BZI9, Q9BZJ1, Q9BZJ2, Q9GZW7, Q9H8F8, Q9NQH5, Q9NYD8 | Gene names | CRNKL1, CRN | |||
|
Domain Architecture |

|
|||||
| Description | Crooked neck-like protein 1 (Crooked neck homolog) (hCrn). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.062279 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.078189 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.076857 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TTC7B_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.099673 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.044517 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.099254 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.063075 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.031348 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.058710 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DTD1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.083649 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TEA8, Q496D1, Q5W184, Q8WXU8, Q9BW67, Q9H464, Q9H474 | Gene names | HARS2, C20orf88 | |||
|
Domain Architecture |

|
|||||
| Description | Probable D-tyrosyl-tRNA(Tyr) deacylase (EC 3.1.-.-). | |||||
|
STK6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.009199 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14965, O60445, O75873, Q9BQD6, Q9UPG5 | Gene names | STK6, AIK, ARK1, AURA, BTAK, STK15 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Serine/threonine kinase 15) (Aurora/IPL1-related kinase 1) (Aurora-related kinase 1) (hARK1) (Aurora-A) (Breast-tumor-amplified kinase). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.058247 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
STK6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.008864 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97477, O35624, Q91YU4 | Gene names | Stk6, Ark1, Aurka, Ayk1, Iak1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Aurora-family kinase 1) (Aurora/IPL1-related kinase 1) (Ipl1- and aurora-related kinase 1) (Aurora-A) (Serine/threonine-protein kinase Ayk1). | |||||
|
CP26A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.024626 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55127, Q9R1F4 | Gene names | Cyp26a1, Cyp26, P450ra | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 26A1 (EC 1.14.-.-) (Retinoic acid-metabolizing cytochrome) (P450RAI) (Retinoic acid 4-hydroxylase). | |||||
|
K1279_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.050116 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPU9, Q5BKR5, Q7TPE0, Q80VQ5, Q8BZE2, Q8CFS7 | Gene names | Kiaa1279 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1279. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.030885 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.090732 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.045378 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TTC8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.082266 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.056701 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DTD1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.072385 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DD18, Q9CRE8, Q9CYL0, Q9D013, Q9D1G4 | Gene names | Hars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable D-tyrosyl-tRNA(Tyr) deacylase (EC 3.1.-.-). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.081622 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
IFIT1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.074458 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09914, Q96QM5 | Gene names | IFIT1, G10P1, IFI56 | |||
|
Domain Architecture |
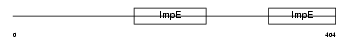
|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.046546 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
KLC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.037592 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.033806 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.050104 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
STIP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.090269 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
CP26A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.021838 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43174 | Gene names | CYP26A1, CYP26, P450RAI1 | |||
|
Domain Architecture |
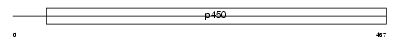
|
|||||
| Description | Cytochrome P450 26A1 (EC 1.14.-.-) (Retinoic acid-metabolizing cytochrome) (P450 retinoic acid-inactivating 1) (P450RAI) (hP450RAI) (Retinoic acid 4-hydroxylase). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.053978 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
P52K_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.070861 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.059835 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
UTY_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.079380 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P79457, O97979 | Gene names | Uty | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein ON the Y chromosome) (Male- specific histocompatibility antigen H-YDB). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.045611 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.042164 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.043910 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.041581 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.039763 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.023675 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
A4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.036934 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.047851 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.045618 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.042179 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.049858 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.051741 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.021620 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TPM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.046832 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
KIF23_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.030760 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.046632 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TPM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.045841 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
WDR19_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.030048 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEZ3, Q8N5B4, Q9H5S0, Q9HCD4 | Gene names | WDR19, KIAA1638 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 19. | |||||
|
A4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.038807 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.029258 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
GGT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.016634 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60928 | Gene names | Ggt1, Ggt, Ggtp | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-glutamyltranspeptidase 1 precursor (EC 2.3.2.2) (Gamma- glutamyltransferase 1) (GGT 1) (CD224 antigen) [Contains: Gamma- glutamyltranspeptidase 1 heavy chain; Gamma-glutamyltranspeptidase 1 light chain]. | |||||
|
KIF11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.023821 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |
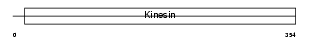
|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.027279 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.044529 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.046322 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RF1M_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.028622 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K126 | Gene names | Mtrf1 | |||
|
Domain Architecture |
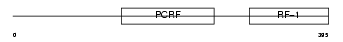
|
|||||
| Description | Peptide chain release factor 1, mitochondrial precursor (MRF-1). | |||||
|
TTC13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.082689 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
TTC7A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.076720 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULT0, Q6PIX4, Q8ND67, Q9BUS3 | Gene names | TTC7A, KIAA1140, TTC7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
TTC8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.065254 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |
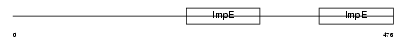
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
AURKB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.005534 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70126, Q61882 | Gene names | Aurkb, Ark2, Stk1, Stk12, Stk5 | |||
|
Domain Architecture |
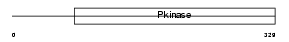
|
|||||
| Description | Serine/threonine-protein kinase 12 (EC 2.7.11.1) (Aurora-B) (Aurora- related kinase 2) (Serine/threonine-protein kinase 5) (STK-1). | |||||
|
CDC23_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.071263 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
FRG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.028155 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97376, Q99M42 | Gene names | Frg1 | |||
|
Domain Architecture |
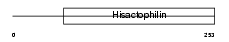
|
|||||
| Description | Protein FRG1 (FSHD region gene 1 protein). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.044013 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
SYCP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.062021 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
TEKT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.038009 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q969V4 | Gene names | TEKT1 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-1. | |||||
|
VPS53_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.024571 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VIR6, Q8WYW3, Q9BRR2, Q9BY02, Q9NV25 | Gene names | VPS53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
VPS53_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.023880 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CCB4, Q8C7U0, Q8CIA1, Q9CX82, Q9D6T7 | Gene names | Vps53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
WDR19_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.021302 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UGF1, Q6ZPK8, Q80VQ6, Q8C794, Q8K3R5 | Gene names | Wdr19, Kiaa1638 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 19. | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.022017 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
BPIL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.022714 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C1E1, Q3TUY4, Q8BVZ0, Q8C1E2, Q9D713, Q9D744 | Gene names | Bpil1 | |||
|
Domain Architecture |

|
|||||
| Description | Bactericidal/permeability-increasing protein-like 1 precursor. | |||||
|
IFIT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.023715 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09913 | Gene names | IFIT2, G10P2, IFI54 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (ISG-54 K). | |||||
|
K0372_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.139350 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.033017 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KI67_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.018661 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.021327 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.022888 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.038225 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.040579 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.041526 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.039952 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
TTC7A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.071042 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BGB2, Q80XT5 | Gene names | Ttc7a, Ttc7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
UTY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.048790 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.031746 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
AN32E_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.023938 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
AN32E_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.022904 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97822, Q3TH89, Q8BPF8, Q8C2L4, Q8C7Q8, Q9CZD2 | Gene names | Anp32e, Cpd1 | |||
|
Domain Architecture |
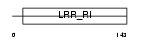
|
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L) (Cerebellar postnatal development protein 1). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.037626 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.033437 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
FANCG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.011324 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQR6 | Gene names | Fancg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein homolog (Protein FACG). | |||||
|
FRG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.022574 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14331 | Gene names | FRG1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein FRG1 (FSHD region gene 1 protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.044824 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.032769 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.040092 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.022120 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LY10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.010671 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.021348 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.004213 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UTX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.052905 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
CDC23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054682 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CDC27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.080490 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.055357 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.054495 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
GEMI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.055920 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.057225 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
TTC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.054116 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
NARG1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q9BXJ9, Q8IWH4, Q8NEV2, Q9H8P6 | Gene names | NARG1, GA19, NATH, TBDN100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Tubedown-1 protein) (Tbdn100) (Gastric cancer antigen Ga19). | |||||
|
NARG1_MOUSE
|
||||||
| NC score | 0.998997 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q80UM3, Q811Z9, Q9JID5 | Gene names | Narg1, Nat1, Tbdn-1, Tubedown | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase 1) (Protein Tubedown-1). | |||||
|
NARGL_MOUSE
|
||||||
| NC score | 0.969679 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
NARGL_HUMAN
|
||||||
| NC score | 0.959732 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.171551 (rank : 5) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
BBS4_HUMAN
|
||||||
| NC score | 0.166736 (rank : 6) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| NC score | 0.165661 (rank : 7) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
IFT88_HUMAN
|
||||||
| NC score | 0.162828 (rank : 8) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.158588 (rank : 9) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
K0372_HUMAN
|
||||||
| NC score | 0.139350 (rank : 10) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
CRNL1_MOUSE
|
||||||
| NC score | 0.127841 (rank : 11) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63154, Q542E8, Q9CQC1 | Gene names | Crnkl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crooked neck-like protein 1 (Crooked neck homolog). | |||||
|
CRNL1_HUMAN
|
||||||
| NC score | 0.122229 (rank : 12) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZJ0, Q5JY64, Q8WYI5, Q9BZI9, Q9BZJ1, Q9BZJ2, Q9GZW7, Q9H8F8, Q9NQH5, Q9NYD8 | Gene names | CRNKL1, CRN | |||
|
Domain Architecture |

|
|||||
| Description | Crooked neck-like protein 1 (Crooked neck homolog) (hCrn). | |||||
|
TTC7B_HUMAN
|
||||||
| NC score | 0.099673 (rank : 13) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.099254 (rank : 14) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.090732 (rank : 15) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.090269 (rank : 16) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
DTD1_HUMAN
|
||||||
| NC score | 0.083649 (rank : 17) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TEA8, Q496D1, Q5W184, Q8WXU8, Q9BW67, Q9H464, Q9H474 | Gene names | HARS2, C20orf88 | |||
|
Domain Architecture |

|
|||||
| Description | Probable D-tyrosyl-tRNA(Tyr) deacylase (EC 3.1.-.-). | |||||
|
TTC13_HUMAN
|
||||||
| NC score | 0.082689 (rank : 18) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
TTC8_HUMAN
|
||||||
| NC score | 0.082266 (rank : 19) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.081622 (rank : 20) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.080490 (rank : 21) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
UTY_MOUSE
|
||||||
| NC score | 0.079380 (rank : 22) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P79457, O97979 | Gene names | Uty | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein ON the Y chromosome) (Male- specific histocompatibility antigen H-YDB). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.079123 (rank : 23) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.078189 (rank : 24) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.076857 (rank : 25) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TTC7A_HUMAN
|
||||||
| NC score | 0.076720 (rank : 26) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULT0, Q6PIX4, Q8ND67, Q9BUS3 | Gene names | TTC7A, KIAA1140, TTC7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
IFIT1_HUMAN
|
||||||
| NC score | 0.074458 (rank : 27) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09914, Q96QM5 | Gene names | IFIT1, G10P1, IFI56 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K). | |||||
|
DTD1_MOUSE
|
||||||
| NC score | 0.072385 (rank : 28) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DD18, Q9CRE8, Q9CYL0, Q9D013, Q9D1G4 | Gene names | Hars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable D-tyrosyl-tRNA(Tyr) deacylase (EC 3.1.-.-). | |||||
|
CDC23_HUMAN
|
||||||
| NC score | 0.071263 (rank : 29) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
TTC7A_MOUSE
|
||||||
| NC score | 0.071042 (rank : 30) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BGB2, Q80XT5 | Gene names | Ttc7a, Ttc7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.070861 (rank : 31) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
TTC8_MOUSE
|
||||||
| NC score | 0.065254 (rank : 32) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.063075 (rank : 33) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.062279 (rank : 34) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
SYCP2_HUMAN
|
||||||
| NC score | 0.062021 (rank : 35) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.059835 (rank : 36) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.058710 (rank : 37) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.058247 (rank : 38) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.057225 (rank : 39) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.056701 (rank : 40) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
GEMI_MOUSE
|
||||||
| NC score | 0.055920 (rank : 41) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.055357 (rank : 42) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CDC23_MOUSE
|
||||||
| NC score | 0.054682 (rank : 43) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.054495 (rank : 44) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.054116 (rank : 45) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.053978 (rank : 46) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
UTX_HUMAN
|
||||||
| NC score | 0.052905 (rank : 47) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.051741 (rank : 48) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
K1279_MOUSE
|
||||||
| NC score | 0.050116 (rank : 49) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPU9, Q5BKR5, Q7TPE0, Q80VQ5, Q8BZE2, Q8CFS7 | Gene names | Kiaa1279 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1279. | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.050104 (rank : 50) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.049858 (rank : 51) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
UTY_HUMAN
|
||||||
| NC score | 0.048790 (rank : 52) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.047851 (rank : 53) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
TPM1_MOUSE
|
||||||
| NC score | 0.046832 (rank : 54) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.046632 (rank : 55) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.046546 (rank : 56) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.046322 (rank : 57) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
TPM1_HUMAN
|
||||||
| NC score | 0.045841 (rank : 58) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.045618 (rank : 59) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.045611 (rank : 60) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.045378 (rank : 61) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.044824 (rank : 62) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.044529 (rank : 63) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.044517 (rank : 64) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.044013 (rank : 65) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.043910 (rank : 66) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.043014 (rank : 67) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.042179 (rank : 68) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.042164 (rank : 69) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.041581 (rank : 70) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.041526 (rank : 71) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.040579 (rank : 72) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.040465 (rank : 73) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.040092 (rank : 74) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.039952 (rank : 75) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.039763 (rank : 76) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.038807 (rank : 77) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.038225 (rank : 78) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
TEKT1_HUMAN
|
||||||
| NC score | 0.038009 (rank : 79) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q969V4 | Gene names | TEKT1 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-1. | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.037626 (rank : 80) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
KLC1_HUMAN
|
||||||
| NC score | 0.037592 (rank : 81) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.036934 (rank : 82) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.033806 (rank : 83) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.033437 (rank : 84) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.033017 (rank : 85) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.032769 (rank : 86) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.031746 (rank : 87) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.031348 (rank : 88) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.030885 (rank : 89) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
KIF23_HUMAN
|
||||||
| NC score | 0.030760 (rank : 90) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
WDR19_HUMAN
|
||||||
| NC score | 0.030048 (rank : 91) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEZ3, Q8N5B4, Q9H5S0, Q9HCD4 | Gene names | WDR19, KIAA1638 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 19. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.029258 (rank : 92) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
RF1M_MOUSE
|
||||||
| NC score | 0.028622 (rank : 93) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K126 | Gene names | Mtrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptide chain release factor 1, mitochondrial precursor (MRF-1). | |||||
|
FRG1_MOUSE
|
||||||
| NC score | 0.028155 (rank : 94) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97376, Q99M42 | Gene names | Frg1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FRG1 (FSHD region gene 1 protein). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.027279 (rank : 95) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
CP26A_MOUSE
|
||||||
| NC score | 0.024626 (rank : 96) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55127, Q9R1F4 | Gene names | Cyp26a1, Cyp26, P450ra | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 26A1 (EC 1.14.-.-) (Retinoic acid-metabolizing cytochrome) (P450RAI) (Retinoic acid 4-hydroxylase). | |||||
|
VPS53_HUMAN
|
||||||
| NC score | 0.024571 (rank : 97) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VIR6, Q8WYW3, Q9BRR2, Q9BY02, Q9NV25 | Gene names | VPS53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
AN32E_HUMAN
|
||||||
| NC score | 0.023938 (rank : 98) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
VPS53_MOUSE
|
||||||
| NC score | 0.023880 (rank : 99) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CCB4, Q8C7U0, Q8CIA1, Q9CX82, Q9D6T7 | Gene names | Vps53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
KIF11_HUMAN
|
||||||
| NC score | 0.023821 (rank : 100) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
IFIT2_HUMAN
|
||||||
| NC score | 0.023715 (rank : 101) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09913 | Gene names | IFIT2, G10P2, IFI54 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (ISG-54 K). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.023675 (rank : 102) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
AN32E_MOUSE
|
||||||
| NC score | 0.022904 (rank : 103) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97822, Q3TH89, Q8BPF8, Q8C2L4, Q8C7Q8, Q9CZD2 | Gene names | Anp32e, Cpd1 | |||
|
Domain Architecture |

|
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L) (Cerebellar postnatal development protein 1). | |||||
|
LSD1_MOUSE
|
||||||
| NC score | 0.022888 (rank : 104) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
BPIL1_MOUSE
|
||||||
| NC score | 0.022714 (rank : 105) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C1E1, Q3TUY4, Q8BVZ0, Q8C1E2, Q9D713, Q9D744 | Gene names | Bpil1 | |||
|
Domain Architecture |

|
|||||
| Description | Bactericidal/permeability-increasing protein-like 1 precursor. | |||||
|
FRG1_HUMAN
|
||||||
| NC score | 0.022574 (rank : 106) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14331 | Gene names | FRG1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FRG1 (FSHD region gene 1 protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.022120 (rank : 107) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.022017 (rank : 108) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
CP26A_HUMAN
|
||||||
| NC score | 0.021838 (rank : 109) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43174 | Gene names | CYP26A1, CYP26, P450RAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 26A1 (EC 1.14.-.-) (Retinoic acid-metabolizing cytochrome) (P450 retinoic acid-inactivating 1) (P450RAI) (hP450RAI) (Retinoic acid 4-hydroxylase). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.021620 (rank : 110) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.021348 (rank : 111) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.021327 (rank : 112) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
WDR19_MOUSE
|
||||||
| NC score | 0.021302 (rank : 113) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UGF1, Q6ZPK8, Q80VQ6, Q8C794, Q8K3R5 | Gene names | Wdr19, Kiaa1638 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 19. | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.018661 (rank : 114) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
GGT1_MOUSE
|
||||||
| NC score | 0.016634 (rank : 115) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60928 | Gene names | Ggt1, Ggt, Ggtp | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-glutamyltranspeptidase 1 precursor (EC 2.3.2.2) (Gamma- glutamyltransferase 1) (GGT 1) (CD224 antigen) [Contains: Gamma- glutamyltranspeptidase 1 heavy chain; Gamma-glutamyltranspeptidase 1 light chain]. | |||||
|
FANCG_MOUSE
|
||||||
| NC score | 0.011324 (rank : 116) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQR6 | Gene names | Fancg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein homolog (Protein FACG). | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.010671 (rank : 117) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
STK6_HUMAN
|
||||||
| NC score | 0.009199 (rank : 118) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14965, O60445, O75873, Q9BQD6, Q9UPG5 | Gene names | STK6, AIK, ARK1, AURA, BTAK, STK15 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Serine/threonine kinase 15) (Aurora/IPL1-related kinase 1) (Aurora-related kinase 1) (hARK1) (Aurora-A) (Breast-tumor-amplified kinase). | |||||
|
STK6_MOUSE
|
||||||
| NC score | 0.008864 (rank : 119) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97477, O35624, Q91YU4 | Gene names | Stk6, Ark1, Aurka, Ayk1, Iak1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Aurora-family kinase 1) (Aurora/IPL1-related kinase 1) (Ipl1- and aurora-related kinase 1) (Aurora-A) (Serine/threonine-protein kinase Ayk1). | |||||
|
AURKB_MOUSE
|
||||||
| NC score | 0.005534 (rank : 120) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70126, Q61882 | Gene names | Aurkb, Ark2, Stk1, Stk12, Stk5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 12 (EC 2.7.11.1) (Aurora-B) (Aurora- related kinase 2) (Serine/threonine-protein kinase 5) (STK-1). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.004213 (rank : 121) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||