Please be patient as the page loads
|
UTY_HUMAN
|
||||||
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UTX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.968410 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
UTY_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
UTY_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.972630 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P79457, O97979 | Gene names | Uty | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein ON the Y chromosome) (Male- specific histocompatibility antigen H-YDB). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 4) | NC score | 0.315739 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
CDC23_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 5) | NC score | 0.357363 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CDC23_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 6) | NC score | 0.356721 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
K0372_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 7) | NC score | 0.300136 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 8) | NC score | 0.212633 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 9) | NC score | 0.208194 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
CDC27_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 10) | NC score | 0.297710 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 11) | NC score | 0.216543 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.210755 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
TTC8_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 13) | NC score | 0.246316 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |
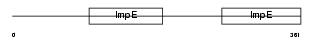
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
IFT88_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.232915 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
TTC8_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 15) | NC score | 0.234090 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |
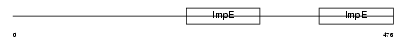
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
IFT88_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.210996 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
TTC6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.187443 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
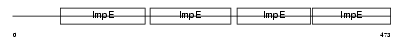
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
PEX5_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.152237 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
TRRAP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.082244 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.027804 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
TTC7A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.135285 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGB2, Q80XT5 | Gene names | Ttc7a, Ttc7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.033587 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.073028 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TTC7A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.124912 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULT0, Q6PIX4, Q8ND67, Q9BUS3 | Gene names | TTC7A, KIAA1140, TTC7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
PEX5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.138330 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.031339 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
TTC12_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.069842 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |
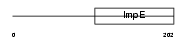
|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.107285 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N0Z6 | Gene names | TTC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
EPC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.027641 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
FANCA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.047499 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JL70, Q9ESU9, Q9ESV3, Q9ET44, Q9JL64, Q9JL65, Q9JL66, Q9JL67, Q9JL68, Q9JL69 | Gene names | Fanca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group A protein homolog (Protein FACA). | |||||
|
KLC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.079241 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.077024 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.067928 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.016110 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NCOA5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.037972 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
BBS10_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.031333 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBI2 | Gene names | Bbs10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bardet-Biedl syndrome 10 protein homolog. | |||||
|
BBS4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.182137 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
CDC16_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.141602 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
PEX5R_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.120089 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
TTC5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.091312 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LG4 | Gene names | Ttc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
KLC3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.061211 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
P66A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.037874 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.000832 (rank : 98) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ACRBP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.025961 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEB7, Q9BY87 | Gene names | ACRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acrosin-binding protein precursor (Proacrosin-binding protein sp32) (Cancer testis antigen OY-TES-1). | |||||
|
APC7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.099745 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
APC7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.099534 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
CDC16_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.133335 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |
|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
IFIT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.049015 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64112, Q62385 | Gene names | Ifit2, Garg39, Ifi54 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (Glucocorticoid- attenuated response gene 39 protein) (GARG-39). | |||||
|
ITF2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.026117 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
MN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.032089 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
AF10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.021741 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
P66A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.030962 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.112285 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
COPE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.022819 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O89079, Q9JM65 | Gene names | Cope, Cope1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coatomer subunit epsilon (Epsilon-coat protein) (Epsilon-COP). | |||||
|
IFIT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.059283 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14879, Q99634, Q9BSK7 | Gene names | IFIT3, IFI60, IFIT4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (IFIT-4) (Interferon-induced 60 kDa protein) (IFI-60K) (ISG-60) (CIG49) (Retinoic acid-induced gene G protein) (RIG-G). | |||||
|
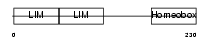
LHX5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.007169 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2C1 | Gene names | LHX5 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
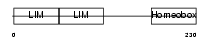
LHX5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.007117 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61375, P50459 | Gene names | Lhx5 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
NARG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.051333 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80UM3, Q811Z9, Q9JID5 | Gene names | Narg1, Nat1, Tbdn-1, Tubedown | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase 1) (Protein Tubedown-1). | |||||
|
NARGL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.068123 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
PEX5R_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.107965 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
VMDL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.012329 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N1M1, Q8N356, Q8NFT9, Q9BR80 | Gene names | VMD2L3 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-4 (Vitelliform macular dystrophy 2-like protein 3). | |||||
|
VWF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.007596 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.012414 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.005739 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
PHF22_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.016828 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
SNIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.017378 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAD8, Q96SP9, Q9H9T7 | Gene names | SNIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
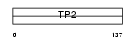
STP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.035677 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05952, Q9NZB0 | Gene names | TNP2 | |||
|
Domain Architecture |
|
|||||
| Description | Nuclear transition protein 2 (TP-2) (TP2). | |||||
|
WDTC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.014549 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.010176 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.012956 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
APBP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.034451 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92624, O95095, Q8WVC9 | Gene names | APPBP2, KIAA0228, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2) (Protein interacting with APP tail 1). | |||||
|
APBP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.034452 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DAX9, Q80U61 | Gene names | Appbp2, Kiaa0228 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2). | |||||
|
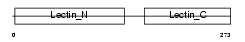
ASGR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.006074 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
DUS10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.009668 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
KLC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.055772 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
NARG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.048790 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXJ9, Q8IWH4, Q8NEV2, Q9H8P6 | Gene names | NARG1, GA19, NATH, TBDN100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Tubedown-1 protein) (Tbdn100) (Gastric cancer antigen Ga19). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.007862 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SNAG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.021104 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99747, Q9BUV1 | Gene names | NAPG, SNAPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-soluble NSF attachment protein (SNAP-gamma) (N-ethylmaleimide- sensitive factor attachment protein, gamma). | |||||
|
SNAG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.020785 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CWZ7, Q3TPT4 | Gene names | Napg, Snapg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-soluble NSF attachment protein (SNAP-gamma) (N-ethylmaleimide- sensitive factor attachment protein, gamma). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.012302 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZNF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | -0.001940 (rank : 99) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17098, Q6PI99 | Gene names | ZNF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 8 (Zinc finger protein HF.18). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.010196 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
DACH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.010015 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
GPC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.007106 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFZ4 | Gene names | Gpc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-3 precursor. | |||||
|
IFIT5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.058940 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13325 | Gene names | IFIT5, RI58 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 5 (IFIT-5) (Retinoic acid- and interferon-inducible 58 kDa protein). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.006714 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MTMR7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.005440 (rank : 97) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
NARGL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.064325 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.008313 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
BBS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.165013 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
KLC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.051413 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.051348 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
NPHP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.054116 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
NPHP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.051304 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
STIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.059506 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.057584 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
TTC13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.145666 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
TTC16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050699 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TTC7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.072552 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
UTY_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
UTY_MOUSE
|
||||||
| NC score | 0.972630 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P79457, O97979 | Gene names | Uty | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein ON the Y chromosome) (Male- specific histocompatibility antigen H-YDB). | |||||
|
UTX_HUMAN
|
||||||
| NC score | 0.968410 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
CDC23_HUMAN
|
||||||
| NC score | 0.357363 (rank : 4) | θ value | 3.41135e-07 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CDC23_MOUSE
|
||||||
| NC score | 0.356721 (rank : 5) | θ value | 3.41135e-07 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.315739 (rank : 6) | θ value | 1.53129e-07 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
K0372_HUMAN
|
||||||
| NC score | 0.300136 (rank : 7) | θ value | 5.44631e-05 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.297710 (rank : 8) | θ value | 0.000270298 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
TTC8_HUMAN
|
||||||
| NC score | 0.246316 (rank : 9) | θ value | 0.00298849 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
TTC8_MOUSE
|
||||||
| NC score | 0.234090 (rank : 10) | θ value | 0.0113563 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.232915 (rank : 11) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.216543 (rank : 12) | θ value | 0.000602161 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.212633 (rank : 13) | θ value | 7.1131e-05 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
IFT88_HUMAN
|
||||||
| NC score | 0.210996 (rank : 14) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.210755 (rank : 15) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.208194 (rank : 16) | θ value | 0.000121331 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.187443 (rank : 17) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
BBS4_MOUSE
|
||||||
| NC score | 0.182137 (rank : 18) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
BBS4_HUMAN
|
||||||
| NC score | 0.165013 (rank : 19) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
PEX5_MOUSE
|
||||||
| NC score | 0.152237 (rank : 20) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
TTC13_HUMAN
|
||||||
| NC score | 0.145666 (rank : 21) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
CDC16_HUMAN
|
||||||
| NC score | 0.141602 (rank : 22) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
PEX5_HUMAN
|
||||||
| NC score | 0.138330 (rank : 23) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
TTC7A_MOUSE
|
||||||
| NC score | 0.135285 (rank : 24) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGB2, Q80XT5 | Gene names | Ttc7a, Ttc7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
CDC16_MOUSE
|
||||||
| NC score | 0.133335 (rank : 25) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
TTC7A_HUMAN
|
||||||
| NC score | 0.124912 (rank : 26) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULT0, Q6PIX4, Q8ND67, Q9BUS3 | Gene names | TTC7A, KIAA1140, TTC7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
PEX5R_MOUSE
|
||||||
| NC score | 0.120089 (rank : 27) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.112285 (rank : 28) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
PEX5R_HUMAN
|
||||||
| NC score | 0.107965 (rank : 29) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
TTC5_HUMAN
|
||||||
| NC score | 0.107285 (rank : 30) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N0Z6 | Gene names | TTC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
APC7_HUMAN
|
||||||
| NC score | 0.099745 (rank : 31) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
APC7_MOUSE
|
||||||
| NC score | 0.099534 (rank : 32) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
TTC5_MOUSE
|
||||||
| NC score | 0.091312 (rank : 33) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LG4 | Gene names | Ttc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
TRRAP_MOUSE
|
||||||
| NC score | 0.082244 (rank : 34) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
KLC2_HUMAN
|
||||||
| NC score | 0.079241 (rank : 35) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC2_MOUSE
|
||||||
| NC score | 0.077024 (rank : 36) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.073028 (rank : 37) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TTC7B_HUMAN
|
||||||
| NC score | 0.072552 (rank : 38) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
TTC12_MOUSE
|
||||||
| NC score | 0.069842 (rank : 39) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
NARGL_MOUSE
|
||||||
| NC score | 0.068123 (rank : 40) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
KLC3_HUMAN
|
||||||
| NC score | 0.067928 (rank : 41) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
NARGL_HUMAN
|
||||||
| NC score | 0.064325 (rank : 42) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
KLC3_MOUSE
|
||||||
| NC score | 0.061211 (rank : 43) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.059506 (rank : 44) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
IFIT3_HUMAN
|
||||||
| NC score | 0.059283 (rank : 45) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14879, Q99634, Q9BSK7 | Gene names | IFIT3, IFI60, IFIT4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (IFIT-4) (Interferon-induced 60 kDa protein) (IFI-60K) (ISG-60) (CIG49) (Retinoic acid-induced gene G protein) (RIG-G). | |||||
|
IFIT5_HUMAN
|
||||||
| NC score | 0.058940 (rank : 46) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13325 | Gene names | IFIT5, RI58 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 5 (IFIT-5) (Retinoic acid- and interferon-inducible 58 kDa protein). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.057584 (rank : 47) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
KLC1_HUMAN
|
||||||
| NC score | 0.055772 (rank : 48) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
NPHP3_HUMAN
|
||||||
| NC score | 0.054116 (rank : 49) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.051413 (rank : 50) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC4_MOUSE
|
||||||
| NC score | 0.051348 (rank : 51) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
NARG1_MOUSE
|
||||||
| NC score | 0.051333 (rank : 52) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80UM3, Q811Z9, Q9JID5 | Gene names | Narg1, Nat1, Tbdn-1, Tubedown | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase 1) (Protein Tubedown-1). | |||||
|
NPHP3_MOUSE
|
||||||
| NC score | 0.051304 (rank : 53) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
TTC16_MOUSE
|
||||||
| NC score | 0.050699 (rank : 54) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
IFIT2_MOUSE
|
||||||
| NC score | 0.049015 (rank : 55) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64112, Q62385 | Gene names | Ifit2, Garg39, Ifi54 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (Glucocorticoid- attenuated response gene 39 protein) (GARG-39). | |||||
|
NARG1_HUMAN
|
||||||
| NC score | 0.048790 (rank : 56) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXJ9, Q8IWH4, Q8NEV2, Q9H8P6 | Gene names | NARG1, GA19, NATH, TBDN100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Tubedown-1 protein) (Tbdn100) (Gastric cancer antigen Ga19). | |||||
|
FANCA_MOUSE
|
||||||
| NC score | 0.047499 (rank : 57) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JL70, Q9ESU9, Q9ESV3, Q9ET44, Q9JL64, Q9JL65, Q9JL66, Q9JL67, Q9JL68, Q9JL69 | Gene names | Fanca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group A protein homolog (Protein FACA). | |||||
|
NCOA5_HUMAN
|
||||||
| NC score | 0.037972 (rank : 58) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
P66A_HUMAN
|
||||||
| NC score | 0.037874 (rank : 59) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
STP2_HUMAN
|
||||||
| NC score | 0.035677 (rank : 60) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05952, Q9NZB0 | Gene names | TNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transition protein 2 (TP-2) (TP2). | |||||
|
APBP2_MOUSE
|
||||||
| NC score | 0.034452 (rank : 61) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DAX9, Q80U61 | Gene names | Appbp2, Kiaa0228 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2). | |||||
|
APBP2_HUMAN
|
||||||
| NC score | 0.034451 (rank : 62) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92624, O95095, Q8WVC9 | Gene names | APPBP2, KIAA0228, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2) (Protein interacting with APP tail 1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.033587 (rank : 63) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.032089 (rank : 64) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.031339 (rank : 65) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
BBS10_MOUSE
|
||||||
| NC score | 0.031333 (rank : 66) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBI2 | Gene names | Bbs10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bardet-Biedl syndrome 10 protein homolog. | |||||
|
P66A_MOUSE
|
||||||
| NC score | 0.030962 (rank : 67) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.027804 (rank : 68) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
EPC2_HUMAN
|
||||||
| NC score | 0.027641 (rank : 69) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
ITF2_HUMAN
|
||||||
| NC score | 0.026117 (rank : 70) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ACRBP_HUMAN
|
||||||
| NC score | 0.025961 (rank : 71) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEB7, Q9BY87 | Gene names | ACRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acrosin-binding protein precursor (Proacrosin-binding protein sp32) (Cancer testis antigen OY-TES-1). | |||||
|
COPE_MOUSE
|
||||||
| NC score | 0.022819 (rank : 72) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O89079, Q9JM65 | Gene names | Cope, Cope1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coatomer subunit epsilon (Epsilon-coat protein) (Epsilon-COP). | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.021741 (rank : 73) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
SNAG_HUMAN
|
||||||
| NC score | 0.021104 (rank : 74) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99747, Q9BUV1 | Gene names | NAPG, SNAPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-soluble NSF attachment protein (SNAP-gamma) (N-ethylmaleimide- sensitive factor attachment protein, gamma). | |||||
|
SNAG_MOUSE
|
||||||
| NC score | 0.020785 (rank : 75) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CWZ7, Q3TPT4 | Gene names | Napg, Snapg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-soluble NSF attachment protein (SNAP-gamma) (N-ethylmaleimide- sensitive factor attachment protein, gamma). | |||||
|
SNIP1_HUMAN
|
||||||
| NC score | 0.017378 (rank : 76) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAD8, Q96SP9, Q9H9T7 | Gene names | SNIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
PHF22_MOUSE
|
||||||
| NC score | 0.016828 (rank : 77) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.016110 (rank : 78) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
WDTC1_HUMAN
|
||||||
| NC score | 0.014549 (rank : 79) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.012956 (rank : 80) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.012414 (rank : 81) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
VMDL3_HUMAN
|
||||||
| NC score | 0.012329 (rank : 82) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N1M1, Q8N356, Q8NFT9, Q9BR80 | Gene names | VMD2L3 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-4 (Vitelliform macular dystrophy 2-like protein 3). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.012302 (rank : 83) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.010196 (rank : 84) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.010176 (rank : 85) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
DACH1_MOUSE
|
||||||
| NC score | 0.010015 (rank : 86) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
DUS10_HUMAN
|
||||||
| NC score | 0.009668 (rank : 87) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.008313 (rank : 88) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.007862 (rank : 89) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.007596 (rank : 90) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
LHX5_HUMAN
|
||||||
| NC score | 0.007169 (rank : 91) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2C1 | Gene names | LHX5 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
LHX5_MOUSE
|
||||||
| NC score | 0.007117 (rank : 92) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61375, P50459 | Gene names | Lhx5 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
GPC3_MOUSE
|
||||||
| NC score | 0.007106 (rank : 93) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFZ4 | Gene names | Gpc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-3 precursor. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.006714 (rank : 94) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ASGR2_HUMAN
|
||||||
| NC score | 0.006074 (rank : 95) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.005739 (rank : 96) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
MTMR7_HUMAN
|
||||||
| NC score | 0.005440 (rank : 97) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.000832 (rank : 98) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ZNF8_HUMAN
|
||||||
| NC score | -0.001940 (rank : 99) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17098, Q6PI99 | Gene names | ZNF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 8 (Zinc finger protein HF.18). | |||||