Please be patient as the page loads
|
NBR1_MOUSE
|
||||||
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NBR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.975755 (rank : 2) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14596, Q13173, Q15026, Q96GB6, Q9NRF7 | Gene names | NBR1, 1A13B, KIAA0049, M17S2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2) (1A1-3B). | |||||
|
NBR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
CF106_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 3) | NC score | 0.353198 (rank : 3) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H6K1, Q5VV77, Q96MG5, Q9BUR9 | Gene names | C6orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf106. | |||||
|
CF106_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 4) | NC score | 0.328075 (rank : 4) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TT38 | Gene names | D17Wsu92e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf106 homolog. | |||||
|
SQSTM_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 5) | NC score | 0.315448 (rank : 5) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
SQSTM_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 6) | NC score | 0.306850 (rank : 6) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
DTNA_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.130121 (rank : 7) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |
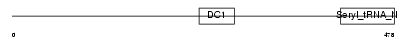
|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
DTNA_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.124841 (rank : 8) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.032066 (rank : 43) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.060834 (rank : 19) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.030725 (rank : 46) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MIB2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.066946 (rank : 16) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
DTNB_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.117398 (rank : 9) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60941, O43782, O60881, O75538, Q9UE14, Q9UE15, Q9UE16 | Gene names | DTNB | |||
|
Domain Architecture |
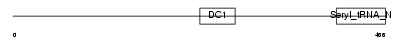
|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B). | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.045002 (rank : 28) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
MIB2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.068154 (rank : 15) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
DTNB_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.113641 (rank : 10) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.044430 (rank : 29) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
MIB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.064593 (rank : 18) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MIB1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.064685 (rank : 17) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.047099 (rank : 24) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.008609 (rank : 105) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.079195 (rank : 13) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.034505 (rank : 38) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.045461 (rank : 26) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
FYB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.077891 (rank : 14) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.028089 (rank : 52) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.033852 (rank : 41) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.032293 (rank : 42) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TTC15_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.040014 (rank : 31) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.051559 (rank : 23) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.031217 (rank : 44) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
MCM8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.019917 (rank : 81) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
OSBP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.018050 (rank : 86) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
SIGL7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.016175 (rank : 93) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y286, Q9NZQ1, Q9UJ86, Q9UJ87, Q9Y502 | Gene names | SIGLEC7, AIRM1 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 7 precursor (Siglec-7) (QA79 membrane protein) (Adhesion inhibitory receptor molecule 1) (AIRM-1) (p75) (D-siglec) (CDw328 antigen). | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.034860 (rank : 37) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
TF7L1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.020000 (rank : 80) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.013252 (rank : 98) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.023695 (rank : 66) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.058424 (rank : 21) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.058811 (rank : 20) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.034449 (rank : 39) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.039259 (rank : 32) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.028036 (rank : 53) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.025442 (rank : 60) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.024263 (rank : 63) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DBNL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.034436 (rank : 40) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.025510 (rank : 58) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.028123 (rank : 50) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SURF6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.053280 (rank : 22) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75683, Q9BRK9, Q9BTZ5, Q9UK24 | Gene names | SURF6, SURF-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.015603 (rank : 95) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.022993 (rank : 68) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DC1I1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.022343 (rank : 71) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88485, O88486 | Gene names | Dync1i1, Dnci1, Dncic1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 1 (Dynein intermediate chain 1, cytosolic) (DH IC-1) (Cytoplasmic dynein intermediate chain 1). | |||||
|
KPCI_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.004846 (rank : 112) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41743, Q8WW06 | Gene names | PRKCI, DXS1179E | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C iota type (EC 2.7.11.13) (nPKC-iota) (Atypical protein kinase C-lambda/iota) (aPKC-lambda/iota) (PRKC-lambda/iota). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.018174 (rank : 85) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.028105 (rank : 51) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.036734 (rank : 35) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.036905 (rank : 34) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CE025_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.031214 (rank : 45) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NDZ2, Q6NXN8, Q6ZTU4, Q8IZ15 | Gene names | C5orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf25. | |||||
|
DPOLZ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.017342 (rank : 89) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.020779 (rank : 77) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
ILF3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.019200 (rank : 82) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
KPCI_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.004603 (rank : 113) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62074 | Gene names | Prkci, Pkcl | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C iota type (EC 2.7.11.13) (nPKC-iota) (Atypical protein kinase C-lambda/iota) (aPKC-lambda/iota). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.004987 (rank : 111) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.043097 (rank : 30) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.026978 (rank : 55) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
REN3A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.045541 (rank : 25) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.024446 (rank : 62) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.021836 (rank : 72) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.009658 (rank : 104) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.038914 (rank : 33) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ZSWM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.080277 (rank : 11) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.008313 (rank : 106) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.011692 (rank : 100) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.025495 (rank : 59) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.021185 (rank : 76) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.022932 (rank : 69) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
LIRB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.007291 (rank : 108) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.021403 (rank : 75) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.007647 (rank : 107) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PRR10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.036220 (rank : 36) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N7Y1 | Gene names | PRR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 10. | |||||
|
SELS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.045202 (rank : 27) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BCZ4, Q921S1, Q9DB55 | Gene names | Sels, H47, Vimp | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein S (VCP-interacting membrane protein) (Minor histocompatibility antigen H47). | |||||
|
ZSWM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.079239 (rank : 12) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.009906 (rank : 103) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
DRG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.029994 (rank : 47) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y295, Q6FGP8, Q8WW69, Q9UGF2 | Gene names | DRG1 | |||
|
Domain Architecture |
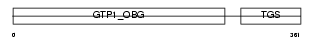
|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1). | |||||
|
DRG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.029982 (rank : 48) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32233 | Gene names | Drg1, Drg, Nedd-3, Nedd3 | |||
|
Domain Architecture |
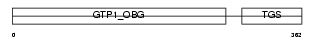
|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1) (Protein NEDD3) (Neural precursor cell expressed developmentally down-regulated protein 3). | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.025664 (rank : 57) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
EOMES_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.006805 (rank : 109) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |
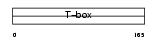
|
|||||
| Description | Eomesodermin homolog. | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.016064 (rank : 94) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
GSH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.003459 (rank : 114) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31316 | Gene names | Gsh2, Gsh-2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GSH-2. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.027318 (rank : 54) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.022531 (rank : 70) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.018267 (rank : 84) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PROX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.023913 (rank : 64) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92786, Q5SW76, Q8TB91 | Gene names | PROX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
PROX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.023845 (rank : 65) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.021740 (rank : 73) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SRA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.021518 (rank : 74) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VJ2, Q8R0S3, Q9CWU7, Q9D973 | Gene names | Sra1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Steroid receptor RNA activator 1 (Steroid receptor RNA activator protein) (SRAP). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.020484 (rank : 79) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
ZBP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.028928 (rank : 49) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H171, Q5JY39, Q9BYW4 | Gene names | ZBP1, C20orf183, DLM1 | |||
|
Domain Architecture |
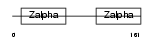
|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
ASPC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.015500 (rank : 96) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZE9, Q7Z6N7, Q8WV59, Q96LS5, Q96M40 | Gene names | ASPSCR1, ASPL, RCC17, TUG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tether containing UBX domain for GLUT4 (Alveolar soft part sarcoma chromosome region candidate 1) (Alveolar soft part sarcoma locus) (Renal papillary cell carcinoma protein 17). | |||||
|
BCL6B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | -0.001722 (rank : 116) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N143, Q6PCB4 | Gene names | BCL6B, BAZF, ZNF62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 6 member B protein (Bcl6-associated zinc finger protein) (Zinc finger protein 62). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.017654 (rank : 87) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.018709 (rank : 83) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.017531 (rank : 88) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.023401 (rank : 67) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
FNDC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.010997 (rank : 101) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BX90, Q2VEY7, Q6ZQ15, Q811D3, Q8BTM3 | Gene names | Fndc3a, D14Ertd453e, Fndc3, Kiaa0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.017151 (rank : 91) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.026183 (rank : 56) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.020571 (rank : 78) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.017324 (rank : 90) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
PARVA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.013346 (rank : 97) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NVD7, Q96C85, Q9HA48 | Gene names | PARVA | |||
|
Domain Architecture |
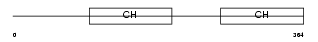
|
|||||
| Description | Alpha-parvin (Calponin-like integrin-linked kinase-binding protein) (CH-ILKBP). | |||||
|
PARVA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.013250 (rank : 99) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPC1, Q9JJ65 | Gene names | Parva, Actp | |||
|
Domain Architecture |
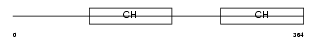
|
|||||
| Description | Alpha-parvin (Actopaxin). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.024817 (rank : 61) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.010941 (rank : 102) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.002396 (rank : 115) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.016408 (rank : 92) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
ZYX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.006348 (rank : 110) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
NBR1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
NBR1_HUMAN
|
||||||
| NC score | 0.975755 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14596, Q13173, Q15026, Q96GB6, Q9NRF7 | Gene names | NBR1, 1A13B, KIAA0049, M17S2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2) (1A1-3B). | |||||
|
CF106_HUMAN
|
||||||
| NC score | 0.353198 (rank : 3) | θ value | 1.99992e-07 (rank : 3) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H6K1, Q5VV77, Q96MG5, Q9BUR9 | Gene names | C6orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf106. | |||||
|
CF106_MOUSE
|
||||||
| NC score | 0.328075 (rank : 4) | θ value | 2.88788e-06 (rank : 4) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TT38 | Gene names | D17Wsu92e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf106 homolog. | |||||
|
SQSTM_HUMAN
|
||||||
| NC score | 0.315448 (rank : 5) | θ value | 8.40245e-06 (rank : 5) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
SQSTM_MOUSE
|
||||||
| NC score | 0.306850 (rank : 6) | θ value | 3.19293e-05 (rank : 6) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
DTNA_MOUSE
|
||||||
| NC score | 0.130121 (rank : 7) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
DTNA_HUMAN
|
||||||
| NC score | 0.124841 (rank : 8) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
DTNB_HUMAN
|
||||||
| NC score | 0.117398 (rank : 9) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60941, O43782, O60881, O75538, Q9UE14, Q9UE15, Q9UE16 | Gene names | DTNB | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B). | |||||
|
DTNB_MOUSE
|
||||||
| NC score | 0.113641 (rank : 10) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
ZSWM2_MOUSE
|
||||||
| NC score | 0.080277 (rank : 11) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
ZSWM2_HUMAN
|
||||||
| NC score | 0.079239 (rank : 12) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.079195 (rank : 13) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.077891 (rank : 14) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
MIB2_MOUSE
|
||||||
| NC score | 0.068154 (rank : 15) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.066946 (rank : 16) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
MIB1_MOUSE
|
||||||
| NC score | 0.064685 (rank : 17) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
MIB1_HUMAN
|
||||||
| NC score | 0.064593 (rank : 18) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.060834 (rank : 19) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.058811 (rank : 20) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.058424 (rank : 21) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
SURF6_HUMAN
|
||||||
| NC score | 0.053280 (rank : 22) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75683, Q9BRK9, Q9BTZ5, Q9UK24 | Gene names | SURF6, SURF-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.051559 (rank : 23) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.047099 (rank : 24) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
REN3A_HUMAN
|
||||||
| NC score | 0.045541 (rank : 25) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.045461 (rank : 26) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
SELS_MOUSE
|
||||||
| NC score | 0.045202 (rank : 27) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BCZ4, Q921S1, Q9DB55 | Gene names | Sels, H47, Vimp | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein S (VCP-interacting membrane protein) (Minor histocompatibility antigen H47). | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.045002 (rank : 28) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.044430 (rank : 29) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.043097 (rank : 30) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TTC15_MOUSE
|
||||||
| NC score | 0.040014 (rank : 31) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.039259 (rank : 32) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.038914 (rank : 33) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.036905 (rank : 34) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.036734 (rank : 35) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
PRR10_HUMAN
|
||||||
| NC score | 0.036220 (rank : 36) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N7Y1 | Gene names | PRR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 10. | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.034860 (rank : 37) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.034505 (rank : 38) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.034449 (rank : 39) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
DBNL_MOUSE
|
||||||
| NC score | 0.034436 (rank : 40) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.033852 (rank : 41) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.032293 (rank : 42) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.032066 (rank : 43) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.031217 (rank : 44) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
CE025_HUMAN
|
||||||
| NC score | 0.031214 (rank : 45) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NDZ2, Q6NXN8, Q6ZTU4, Q8IZ15 | Gene names | C5orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf25. | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.030725 (rank : 46) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
DRG1_HUMAN
|
||||||
| NC score | 0.029994 (rank : 47) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y295, Q6FGP8, Q8WW69, Q9UGF2 | Gene names | DRG1 | |||
|
Domain Architecture |

|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1). | |||||
|
DRG1_MOUSE
|
||||||
| NC score | 0.029982 (rank : 48) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32233 | Gene names | Drg1, Drg, Nedd-3, Nedd3 | |||
|
Domain Architecture |

|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1) (Protein NEDD3) (Neural precursor cell expressed developmentally down-regulated protein 3). | |||||
|
ZBP1_HUMAN
|
||||||
| NC score | 0.028928 (rank : 49) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H171, Q5JY39, Q9BYW4 | Gene names | ZBP1, C20orf183, DLM1 | |||
|
Domain Architecture |

|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.028123 (rank : 50) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.028105 (rank : 51) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.028089 (rank : 52) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.028036 (rank : 53) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.027318 (rank : 54) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.026978 (rank : 55) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.026183 (rank : 56) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.025664 (rank : 57) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.025510 (rank : 58) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.025495 (rank : 59) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.025442 (rank : 60) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.024817 (rank : 61) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.024446 (rank : 62) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.024263 (rank : 63) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
PROX1_HUMAN
|
||||||
| NC score | 0.023913 (rank : 64) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92786, Q5SW76, Q8TB91 | Gene names | PROX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
PROX1_MOUSE
|
||||||
| NC score | 0.023845 (rank : 65) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.023695 (rank : 66) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.023401 (rank : 67) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.022993 (rank : 68) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.022932 (rank : 69) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.022531 (rank : 70) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
DC1I1_MOUSE
|
||||||
| NC score | 0.022343 (rank : 71) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88485, O88486 | Gene names | Dync1i1, Dnci1, Dncic1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 1 (Dynein intermediate chain 1, cytosolic) (DH IC-1) (Cytoplasmic dynein intermediate chain 1). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.021836 (rank : 72) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.021740 (rank : 73) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SRA1_MOUSE
|
||||||
| NC score | 0.021518 (rank : 74) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VJ2, Q8R0S3, Q9CWU7, Q9D973 | Gene names | Sra1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Steroid receptor RNA activator 1 (Steroid receptor RNA activator protein) (SRAP). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.021403 (rank : 75) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.021185 (rank : 76) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.020779 (rank : 77) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.020571 (rank : 78) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.020484 (rank : 79) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
TF7L1_HUMAN
|
||||||
| NC score | 0.020000 (rank : 80) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
MCM8_MOUSE
|
||||||
| NC score | 0.019917 (rank : 81) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
ILF3_MOUSE
|
||||||
| NC score | 0.019200 (rank : 82) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.018709 (rank : 83) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.018267 (rank : 84) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.018174 (rank : 85) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
OSBP2_HUMAN
|
||||||
| NC score | 0.018050 (rank : 86) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.017654 (rank : 87) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.017531 (rank : 88) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
DPOLZ_MOUSE
|
||||||
| NC score | 0.017342 (rank : 89) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.017324 (rank : 90) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.017151 (rank : 91) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.016408 (rank : 92) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
SIGL7_HUMAN
|
||||||
| NC score | 0.016175 (rank : 93) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y286, Q9NZQ1, Q9UJ86, Q9UJ87, Q9Y502 | Gene names | SIGLEC7, AIRM1 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 7 precursor (Siglec-7) (QA79 membrane protein) (Adhesion inhibitory receptor molecule 1) (AIRM-1) (p75) (D-siglec) (CDw328 antigen). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.016064 (rank : 94) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.015603 (rank : 95) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
ASPC1_HUMAN
|
||||||
| NC score | 0.015500 (rank : 96) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZE9, Q7Z6N7, Q8WV59, Q96LS5, Q96M40 | Gene names | ASPSCR1, ASPL, RCC17, TUG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tether containing UBX domain for GLUT4 (Alveolar soft part sarcoma chromosome region candidate 1) (Alveolar soft part sarcoma locus) (Renal papillary cell carcinoma protein 17). | |||||
|
PARVA_HUMAN
|
||||||
| NC score | 0.013346 (rank : 97) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NVD7, Q96C85, Q9HA48 | Gene names | PARVA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-parvin (Calponin-like integrin-linked kinase-binding protein) (CH-ILKBP). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.013252 (rank : 98) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
PARVA_MOUSE
|
||||||
| NC score | 0.013250 (rank : 99) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPC1, Q9JJ65 | Gene names | Parva, Actp | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-parvin (Actopaxin). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.011692 (rank : 100) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
FNDC3_MOUSE
|
||||||
| NC score | 0.010997 (rank : 101) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BX90, Q2VEY7, Q6ZQ15, Q811D3, Q8BTM3 | Gene names | Fndc3a, D14Ertd453e, Fndc3, Kiaa0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.010941 (rank : 102) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.009906 (rank : 103) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.009658 (rank : 104) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.008609 (rank : 105) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.008313 (rank : 106) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.007647 (rank : 107) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
LIRB3_HUMAN
|
||||||
| NC score | 0.007291 (rank : 108) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
EOMES_HUMAN
|
||||||
| NC score | 0.006805 (rank : 109) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
ZYX_HUMAN
|
||||||
| NC score | 0.006348 (rank : 110) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.004987 (rank : 111) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
KPCI_HUMAN
|
||||||
| NC score | 0.004846 (rank : 112) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41743, Q8WW06 | Gene names | PRKCI, DXS1179E | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C iota type (EC 2.7.11.13) (nPKC-iota) (Atypical protein kinase C-lambda/iota) (aPKC-lambda/iota) (PRKC-lambda/iota). | |||||
|
KPCI_MOUSE
|
||||||
| NC score | 0.004603 (rank : 113) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62074 | Gene names | Prkci, Pkcl | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C iota type (EC 2.7.11.13) (nPKC-iota) (Atypical protein kinase C-lambda/iota) (aPKC-lambda/iota). | |||||
|
GSH2_MOUSE
|
||||||
| NC score | 0.003459 (rank : 114) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31316 | Gene names | Gsh2, Gsh-2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GSH-2. | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.002396 (rank : 115) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
BCL6B_HUMAN
|
||||||
| NC score | -0.001722 (rank : 116) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N143, Q6PCB4 | Gene names | BCL6B, BAZF, ZNF62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 6 member B protein (Bcl6-associated zinc finger protein) (Zinc finger protein 62). | |||||