Please be patient as the page loads
|
PARC_MOUSE
|
||||||
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CUL7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.811550 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.986191 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
PARC_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
ARI2_HUMAN
|
||||||
| θ value | 2.19075e-38 (rank : 4) | NC score | 0.688157 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95376, Q9HBZ6, Q9UEM9 | Gene names | ARIH2, ARI2, TRIAD1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-2 homolog (ARI-2) (Triad1 protein). | |||||
|
ARI2_MOUSE
|
||||||
| θ value | 2.86122e-38 (rank : 5) | NC score | 0.687702 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z1K6 | Gene names | Arih2, Ari2, Triad1, Uip48 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-2 homolog (ARI-2) (Triad1 protein) (UbcM4-interacting protein 48). | |||||
|
ARI1_HUMAN
|
||||||
| θ value | 1.79631e-32 (rank : 6) | NC score | 0.672716 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y4X5, O76026, Q9H3T6, Q9UEN0, Q9UP39 | Gene names | ARIH1, ARI, UBCH7BP | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein) (HHARI) (H7-AP2) (MOP-6). | |||||
|
ARI1_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 7) | NC score | 0.672586 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z1K5, Q6PF92 | Gene names | Arih1, Ari, Ubch7bp, Uip77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein 77). | |||||
|
HECD3_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 8) | NC score | 0.363711 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 9) | NC score | 0.363515 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 10) | NC score | 0.267482 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 11) | NC score | 0.260948 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
RNF14_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 12) | NC score | 0.583514 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JI90, Q9D0L2, Q9D6N2, Q9D6Z8, Q9JI89 | Gene names | Rnf14, Ara54, Triad2 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 14 (Androgen receptor-associated protein 54) (Triad2 protein). | |||||
|
RNF14_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 13) | NC score | 0.581319 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UBS8, O94793, Q6IBV0 | Gene names | RNF14, ARA54 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 14 (Androgen receptor-associated protein 54) (Triad2 protein) (HFB30). | |||||
|
IBRD3_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 14) | NC score | 0.552918 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZMZ0, Q6P6A4 | Gene names | IBRDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IBR domain-containing protein 3. | |||||
|
RNF19_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 15) | NC score | 0.521893 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NV58, Q9H5H9, Q9H8M8, Q9UFG0, Q9UFX6, Q9Y4Y1 | Gene names | RNF19 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (Dorfin) (Double ring-finger protein) (p38 protein). | |||||
|
RNF19_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 16) | NC score | 0.522893 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50636, Q9QUJ5 | Gene names | Rnf19, Geg-154, Xybp | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (XY body protein) (XYbp) (Gametogenesis expressed protein GEG-154) (UBCM4-interacting protein 117) (UIP117). | |||||
|
UB7I4_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 17) | NC score | 0.562090 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q925F3, Q6A0B4 | Gene names | Rnf144, Kiaa0161, Ubce7ip4, Uip4 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
UB7I4_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 18) | NC score | 0.558311 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P50876 | Gene names | RNF144, KIAA0161, UBCE7IP4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
IBRD2_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 19) | NC score | 0.542338 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BKD6, Q8BG97 | Gene names | Ibrdc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase IBRDC2 (EC 6.3.2.-) (IBR domain-containing protein 2). | |||||
|
PRKN2_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 20) | NC score | 0.494143 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVS6, Q9ES22, Q9ES23 | Gene names | Park2, Prkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN). | |||||
|
PRKN2_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 21) | NC score | 0.492443 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
RNF31_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 22) | NC score | 0.429151 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
UB7I3_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 23) | NC score | 0.445833 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BYM8, O95623, Q96BS3, Q9BYM9 | Gene names | RBCK1, C20orf18, RNF54, UBCE7IP3, XAP4 | |||
|
Domain Architecture |
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (Hepatitis B virus X- associated protein 4) (HBV-associated factor 4) (RING finger protein 54). | |||||
|
UB7I3_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 24) | NC score | 0.447876 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
RNF31_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 25) | NC score | 0.424143 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
IBRD2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 26) | NC score | 0.528466 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z419, Q6P4Q0, Q8N3R7, Q9BX38 | Gene names | IBRDC2, P53RFP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase IBRDC2 (EC 6.3.2.-) (IBR domain-containing protein 2) (p53-inducible RING finger protein). | |||||
|
IBRD1_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 27) | NC score | 0.526432 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TC41, Q9BX48 | Gene names | IBRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IBR domain-containing protein 1. | |||||
|
CUL4A_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 28) | NC score | 0.097661 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |
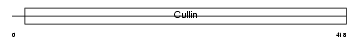
|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
APC10_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 29) | NC score | 0.289799 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UM13, Q2V500, Q9UG51, Q9Y5R0 | Gene names | ANAPC10, APC10 | |||
|
Domain Architecture |
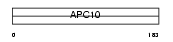
|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
APC10_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 30) | NC score | 0.287927 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2H6, Q9CW24 | Gene names | Anapc10, Apc10 | |||
|
Domain Architecture |
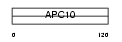
|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
CUL4A_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 31) | NC score | 0.096776 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4B_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 32) | NC score | 0.098267 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
PHLB3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.047120 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NSJ2, Q8N7Z4 | Gene names | PHLDB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 3. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 34) | NC score | 0.013883 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
ABLM1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 35) | NC score | 0.026443 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
ABLM1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.022744 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
FBN3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.021020 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
TRIM9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.050320 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C026, Q92557, Q96D24, Q96NI4, Q9C025, Q9C027 | Gene names | TRIM9, KIAA0282, RNF91 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9 (RING finger protein 91). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.041377 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.053439 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
PDE9A_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.034391 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.033504 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.026066 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CD177_HUMAN
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.057780 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N6Q3, Q711Q2, Q8NCV9, Q96QH1, Q9HDA5 | Gene names | CD177, NB1, PRV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD177 antigen precursor (Polycythemia rubra vera protein 1) (PRV-1) (NB1 glycoprotein) (NB1 GP) (Human neutrophil alloantigen 2a) (HNA- 2a). | |||||
|
PEX10_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.067321 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60683, Q5T095, Q9BW90 | Gene names | PEX10, RNF69 | |||
|
Domain Architecture |
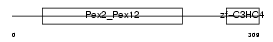
|
|||||
| Description | Peroxisome assembly protein 10 (Peroxin-10) (Peroxisome biogenesis factor 10) (RING finger protein 69). | |||||
|
PRA12_HUMAN
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.015518 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95522 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member DJ1198H6.2. | |||||
|
ERF_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.020756 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.020832 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.022451 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.024338 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PDE9A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.030997 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.002609 (rank : 129) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.017423 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.017149 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.020094 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TRIM9_MOUSE
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.044797 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C7M3, Q6ZQE5, Q80WT6, Q8C7Z4, Q8CC32, Q8CEG2, Q99PQ3 | Gene names | Trim9, Kiaa0282 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9. | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.289986 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.014843 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.021844 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
UB7I1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.292388 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.014064 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.010912 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.065649 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
TOPRS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.066407 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
ADA18_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.011644 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R157, Q60621 | Gene names | Adam18, Adam27, Dtgn3, Tmdc3 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 18 precursor (A disintegrin and metalloproteinase domain 18) (Transmembrane metalloproteinase-like, disintegrin-like, and cysteine- rich protein III) (tMDC III) (ADAM 27). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.011608 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.004659 (rank : 125) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
SAMD4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.028576 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBY1 | Gene names | Samd4 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 4. | |||||
|
TRI37_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.023402 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PCX9, Q8CHC5 | Gene names | Trim37, Kiaa0898 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37. | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.045833 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.043702 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.010385 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
RNF5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.046491 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35445 | Gene names | Rnf5, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (RING finger protein 5). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.003434 (rank : 126) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
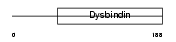
DTBP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.019627 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EV8, Q96NV2, Q9H0U2, Q9H3J5 | Gene names | DTNBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein). | |||||
|
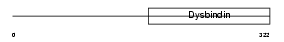
DTBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.019783 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91WZ8, Q80ZN4 | Gene names | Dtnbp1 | |||
|
Domain Architecture |
|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein homolog). | |||||
|
FBN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.012114 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FEZ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.013436 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99689, O00679, O00728, Q6IBI7 | Gene names | FEZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
SAMD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.025398 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU9 | Gene names | SAMD4, KIAA1053 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 4. | |||||
|
BSCL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.014995 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.013313 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
CF049_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.017306 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2TBC4, Q5T3D4, Q9NSV1 | Gene names | C6orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf49. | |||||
|
FLT3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | -0.001891 (rank : 134) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00342 | Gene names | Flt3, Flk-2, Flt-3 | |||
|
Domain Architecture |

|
|||||
| Description | FL cytokine receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor flk-2) (Fetal liver kinase 2) (Tyrosine-protein kinase FLT3) (CD135 antigen). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.030318 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.010764 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
RO52_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.007446 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19474, Q96RF8 | Gene names | TRIM21, RNF81, RO52, SSA1 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa Ro protein (Sjoegren syndrome type A antigen) (SS-A) (Ro(SS-A)) (52 kDa ribonucleoprotein autoantigen Ro/SS-A) (Tripartite motif- containing protein 21) (RING finger protein 81). | |||||
|
TE2IP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.014400 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYB0, Q8WYZ3, Q9NWR2 | Gene names | TERF2IP, RAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1) (hRap1). | |||||
|
DMXL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.006225 (rank : 122) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
GOLI_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.034149 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86XS8, Q641P9, Q6P177, Q7L2U2, Q9P0J9 | Gene names | RNF130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130). | |||||
|
GOLI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.026002 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VEM1, Q80VL7, Q9QZQ6 | Gene names | Rnf130, G1rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130) (G1-related zinc finger protein). | |||||
|
PLXA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.006055 (rank : 123) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.002601 (rank : 130) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
RNF5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.042727 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99942, Q9UMQ2 | Gene names | RNF5, G16, NG2, RMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (HsRma1) (RING finger protein 5) (Protein G16). | |||||
|
FBXL7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.005282 (rank : 124) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJT9, O94926 | Gene names | FBXL7, FBL6, FBL7, KIAA0840 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 7 (F-box and leucine-rich repeat protein 7) (F-box protein FBL6/FBL7). | |||||
|
GRID1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.002656 (rank : 128) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.002998 (rank : 127) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.026355 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.039918 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.039779 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | -0.001527 (rank : 133) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.007216 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RAG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.009519 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
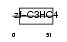
RN152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.044008 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8N0 | Gene names | RNF152 | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 152. | |||||
|
RN152_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.044020 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BG47 | Gene names | Rnf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 152. | |||||
|
RN185_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.041719 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96GF1, Q8N900 | Gene names | RNF185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
RN185_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.041160 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91YT2, Q6ZWS3 | Gene names | Rnf185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | -0.001355 (rank : 132) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.000553 (rank : 131) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.011983 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TRI27_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.009516 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14373, Q5ST26, Q6LA73, Q6NXR9, Q9BZY6, Q9UJL3 | Gene names | TRIM27, RFP, RNF76 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein RFP (Ret finger protein) (Tripartite motif- containing protein 27) (RING finger protein 76). | |||||
|
TRI27_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.008877 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62158, Q62157, Q8C2Q5, Q99LK1 | Gene names | Trim27, Rfp | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein RFP (Ret finger protein) (Tripartite motif- containing protein 27). | |||||
|
TRI36_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.019647 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80WG7, Q7TNM1 | Gene names | Trim36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Haprin) (Acrosome RBCC protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | -0.003179 (rank : 135) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.010956 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CUL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.060622 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.060617 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.055196 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.055230 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.055161 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.055493 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.050762 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.051098 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HERC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.054948 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HERC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.056205 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.055206 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.055054 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
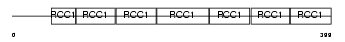
RCC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050808 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
RCC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.051142 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
RCC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.050702 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
SRGEF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.051013 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
SRGEF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.051039 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.057109 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.052274 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054139 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.053806 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
PARC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.986191 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.811550 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
ARI2_HUMAN
|
||||||
| NC score | 0.688157 (rank : 4) | θ value | 2.19075e-38 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95376, Q9HBZ6, Q9UEM9 | Gene names | ARIH2, ARI2, TRIAD1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-2 homolog (ARI-2) (Triad1 protein). | |||||
|
ARI2_MOUSE
|
||||||
| NC score | 0.687702 (rank : 5) | θ value | 2.86122e-38 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z1K6 | Gene names | Arih2, Ari2, Triad1, Uip48 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-2 homolog (ARI-2) (Triad1 protein) (UbcM4-interacting protein 48). | |||||
|
ARI1_HUMAN
|
||||||
| NC score | 0.672716 (rank : 6) | θ value | 1.79631e-32 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y4X5, O76026, Q9H3T6, Q9UEN0, Q9UP39 | Gene names | ARIH1, ARI, UBCH7BP | |||
|
Domain Architecture |

|
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein) (HHARI) (H7-AP2) (MOP-6). | |||||
|
ARI1_MOUSE
|
||||||
| NC score | 0.672586 (rank : 7) | θ value | 1.79631e-32 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z1K5, Q6PF92 | Gene names | Arih1, Ari, Ubch7bp, Uip77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2- binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein 77). | |||||
|
RNF14_MOUSE
|
||||||
| NC score | 0.583514 (rank : 8) | θ value | 2.13179e-17 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JI90, Q9D0L2, Q9D6N2, Q9D6Z8, Q9JI89 | Gene names | Rnf14, Ara54, Triad2 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 14 (Androgen receptor-associated protein 54) (Triad2 protein). | |||||
|
RNF14_HUMAN
|
||||||
| NC score | 0.581319 (rank : 9) | θ value | 1.058e-16 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UBS8, O94793, Q6IBV0 | Gene names | RNF14, ARA54 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 14 (Androgen receptor-associated protein 54) (Triad2 protein) (HFB30). | |||||
|
UB7I4_MOUSE
|
||||||
| NC score | 0.562090 (rank : 10) | θ value | 4.91457e-14 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q925F3, Q6A0B4 | Gene names | Rnf144, Kiaa0161, Ubce7ip4, Uip4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
UB7I4_HUMAN
|
||||||
| NC score | 0.558311 (rank : 11) | θ value | 2.43908e-13 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P50876 | Gene names | RNF144, KIAA0161, UBCE7IP4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
IBRD3_HUMAN
|
||||||
| NC score | 0.552918 (rank : 12) | θ value | 1.68911e-14 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZMZ0, Q6P6A4 | Gene names | IBRDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IBR domain-containing protein 3. | |||||
|
IBRD2_MOUSE
|
||||||
| NC score | 0.542338 (rank : 13) | θ value | 2.28291e-11 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BKD6, Q8BG97 | Gene names | Ibrdc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase IBRDC2 (EC 6.3.2.-) (IBR domain-containing protein 2). | |||||
|
IBRD2_HUMAN
|
||||||
| NC score | 0.528466 (rank : 14) | θ value | 1.38499e-08 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z419, Q6P4Q0, Q8N3R7, Q9BX38 | Gene names | IBRDC2, P53RFP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase IBRDC2 (EC 6.3.2.-) (IBR domain-containing protein 2) (p53-inducible RING finger protein). | |||||
|
IBRD1_HUMAN
|
||||||
| NC score | 0.526432 (rank : 15) | θ value | 2.61198e-07 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TC41, Q9BX48 | Gene names | IBRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IBR domain-containing protein 1. | |||||
|
RNF19_MOUSE
|
||||||
| NC score | 0.522893 (rank : 16) | θ value | 4.91457e-14 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50636, Q9QUJ5 | Gene names | Rnf19, Geg-154, Xybp | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (XY body protein) (XYbp) (Gametogenesis expressed protein GEG-154) (UBCM4-interacting protein 117) (UIP117). | |||||
|
RNF19_HUMAN
|
||||||
| NC score | 0.521893 (rank : 17) | θ value | 4.91457e-14 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NV58, Q9H5H9, Q9H8M8, Q9UFG0, Q9UFX6, Q9Y4Y1 | Gene names | RNF19 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 19 (Dorfin) (Double ring-finger protein) (p38 protein). | |||||
|
PRKN2_MOUSE
|
||||||
| NC score | 0.494143 (rank : 18) | θ value | 3.89403e-11 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVS6, Q9ES22, Q9ES23 | Gene names | Park2, Prkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN). | |||||
|
PRKN2_HUMAN
|
||||||
| NC score | 0.492443 (rank : 19) | θ value | 1.9326e-10 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
UB7I3_MOUSE
|
||||||
| NC score | 0.447876 (rank : 20) | θ value | 5.62301e-10 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
UB7I3_HUMAN
|
||||||
| NC score | 0.445833 (rank : 21) | θ value | 5.62301e-10 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BYM8, O95623, Q96BS3, Q9BYM9 | Gene names | RBCK1, C20orf18, RNF54, UBCE7IP3, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (Hepatitis B virus X- associated protein 4) (HBV-associated factor 4) (RING finger protein 54). | |||||
|
RNF31_HUMAN
|
||||||
| NC score | 0.429151 (rank : 22) | θ value | 2.52405e-10 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
RNF31_MOUSE
|
||||||
| NC score | 0.424143 (rank : 23) | θ value | 1.63604e-09 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.363711 (rank : 24) | θ value | 1.74391e-19 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.363515 (rank : 25) | θ value | 1.74391e-19 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
UB7I1_HUMAN
|
||||||
| NC score | 0.292388 (rank : 26) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.289986 (rank : 27) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
APC10_HUMAN
|
||||||
| NC score | 0.289799 (rank : 28) | θ value | 0.000270298 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UM13, Q2V500, Q9UG51, Q9Y5R0 | Gene names | ANAPC10, APC10 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
APC10_MOUSE
|
||||||
| NC score | 0.287927 (rank : 29) | θ value | 0.000270298 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2H6, Q9CW24 | Gene names | Anapc10, Apc10 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 10 (APC10) (Cyclosome subunit 10). | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.267482 (rank : 30) | θ value | 1.74391e-19 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.260948 (rank : 31) | θ value | 1.24977e-17 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
CUL4B_HUMAN
|
||||||
| NC score | 0.098267 (rank : 32) | θ value | 0.00228821 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CUL4A_HUMAN
|
||||||
| NC score | 0.097661 (rank : 33) | θ value | 0.00020696 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4A_MOUSE
|
||||||
| NC score | 0.096776 (rank : 34) | θ value | 0.00102713 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
PEX10_HUMAN
|
||||||
| NC score | 0.067321 (rank : 35) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60683, Q5T095, Q9BW90 | Gene names | PEX10, RNF69 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly protein 10 (Peroxin-10) (Peroxisome biogenesis factor 10) (RING finger protein 69). | |||||
|
TOPRS_MOUSE
|
||||||
| NC score | 0.066407 (rank : 36) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.065649 (rank : 37) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
CUL1_HUMAN
|
||||||
| NC score | 0.060622 (rank : 38) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| NC score | 0.060617 (rank : 39) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CD177_HUMAN
|
||||||
| NC score | 0.057780 (rank : 40) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N6Q3, Q711Q2, Q8NCV9, Q96QH1, Q9HDA5 | Gene names | CD177, NB1, PRV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD177 antigen precursor (Polycythemia rubra vera protein 1) (PRV-1) (NB1 glycoprotein) (NB1 GP) (Human neutrophil alloantigen 2a) (HNA- 2a). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.057109 (rank : 41) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.056205 (rank : 42) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.055493 (rank : 43) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
CUL3_MOUSE
|
||||||
| NC score | 0.055230 (rank : 44) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.055206 (rank : 45) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
CUL3_HUMAN
|
||||||
| NC score | 0.055196 (rank : 46) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.055161 (rank : 47) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.055054 (rank : 48) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.054948 (rank : 49) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.054139 (rank : 50) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.053806 (rank : 51) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.053439 (rank : 52) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.052274 (rank : 53) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
RCC2_HUMAN
|
||||||
| NC score | 0.051142 (rank : 54) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.051098 (rank : 55) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
SRGEF_MOUSE
|
||||||
| NC score | 0.051039 (rank : 56) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
SRGEF_HUMAN
|
||||||
| NC score | 0.051013 (rank : 57) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
RCC1_HUMAN
|
||||||
| NC score | 0.050808 (rank : 58) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.050762 (rank : 59) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
RCC2_MOUSE
|
||||||
| NC score | 0.050702 (rank : 60) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
TRIM9_HUMAN
|
||||||
| NC score | 0.050320 (rank : 61) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C026, Q92557, Q96D24, Q96NI4, Q9C025, Q9C027 | Gene names | TRIM9, KIAA0282, RNF91 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9 (RING finger protein 91). | |||||
|
PHLB3_HUMAN
|
||||||
| NC score | 0.047120 (rank : 62) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NSJ2, Q8N7Z4 | Gene names | PHLDB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 3. | |||||
|
RNF5_MOUSE
|
||||||
| NC score | 0.046491 (rank : 63) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35445 | Gene names | Rnf5, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (RING finger protein 5). | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.045833 (rank : 64) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
TRIM9_MOUSE
|
||||||
| NC score | 0.044797 (rank : 65) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C7M3, Q6ZQE5, Q80WT6, Q8C7Z4, Q8CC32, Q8CEG2, Q99PQ3 | Gene names | Trim9, Kiaa0282 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9. | |||||
|
RN152_MOUSE
|
||||||
| NC score | 0.044020 (rank : 66) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BG47 | Gene names | Rnf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 152. | |||||
|
RN152_HUMAN
|
||||||
| NC score | 0.044008 (rank : 67) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8N0 | Gene names | RNF152 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 152. | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.043702 (rank : 68) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
RNF5_HUMAN
|
||||||
| NC score | 0.042727 (rank : 69) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99942, Q9UMQ2 | Gene names | RNF5, G16, NG2, RMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (HsRma1) (RING finger protein 5) (Protein G16). | |||||
|
RN185_HUMAN
|
||||||
| NC score | 0.041719 (rank : 70) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96GF1, Q8N900 | Gene names | RNF185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.041377 (rank : 71) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
RN185_MOUSE
|
||||||
| NC score | 0.041160 (rank : 72) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91YT2, Q6ZWS3 | Gene names | Rnf185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.039918 (rank : 73) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.039779 (rank : 74) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PDE9A_MOUSE
|
||||||
| NC score | 0.034391 (rank : 75) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
GOLI_HUMAN
|
||||||
| NC score | 0.034149 (rank : 76) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86XS8, Q641P9, Q6P177, Q7L2U2, Q9P0J9 | Gene names | RNF130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.033504 (rank : 77) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PDE9A_HUMAN
|
||||||
| NC score | 0.030997 (rank : 78) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.030318 (rank : 79) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
SAMD4_MOUSE
|
||||||
| NC score | 0.028576 (rank : 80) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBY1 | Gene names | Samd4 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 4. | |||||
|
ABLM1_MOUSE
|
||||||
| NC score | 0.026443 (rank : 81) | θ value | 0.0736092 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.026355 (rank : 82) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.026066 (rank : 83) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
GOLI_MOUSE
|
||||||
| NC score | 0.026002 (rank : 84) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VEM1, Q80VL7, Q9QZQ6 | Gene names | Rnf130, G1rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130) (G1-related zinc finger protein). | |||||
|
SAMD4_HUMAN
|
||||||
| NC score | 0.025398 (rank : 85) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU9 | Gene names | SAMD4, KIAA1053 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 4. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.024338 (rank : 86) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TRI37_MOUSE
|
||||||
| NC score | 0.023402 (rank : 87) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PCX9, Q8CHC5 | Gene names | Trim37, Kiaa0898 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37. | |||||
|
ABLM1_HUMAN
|
||||||
| NC score | 0.022744 (rank : 88) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.022451 (rank : 89) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.021844 (rank : 90) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.021020 (rank : 91) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.020832 (rank : 92) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.020756 (rank : 93) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.020094 (rank : 94) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DTBP1_MOUSE
|
||||||
| NC score | 0.019783 (rank : 95) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91WZ8, Q80ZN4 | Gene names | Dtnbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein homolog). | |||||
|
TRI36_MOUSE
|
||||||
| NC score | 0.019647 (rank : 96) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80WG7, Q7TNM1 | Gene names | Trim36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Haprin) (Acrosome RBCC protein). | |||||
|
DTBP1_HUMAN
|
||||||
| NC score | 0.019627 (rank : 97) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EV8, Q96NV2, Q9H0U2, Q9H3J5 | Gene names | DTNBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.017423 (rank : 98) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
CF049_HUMAN
|
||||||
| NC score | 0.017306 (rank : 99) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2TBC4, Q5T3D4, Q9NSV1 | Gene names | C6orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf49. | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.017149 (rank : 100) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
PRA12_HUMAN
|
||||||
| NC score | 0.015518 (rank : 101) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95522 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRAME family member DJ1198H6.2. | |||||
|
BSCL2_MOUSE
|
||||||
| NC score | 0.014995 (rank : 102) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.014843 (rank : 103) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
TE2IP_HUMAN
|
||||||
| NC score | 0.014400 (rank : 104) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYB0, Q8WYZ3, Q9NWR2 | Gene names | TERF2IP, RAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1) (hRap1). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.014064 (rank : 105) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.013883 (rank : 106) | θ value | 0.0563607 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
FEZ1_HUMAN
|
||||||
| NC score | 0.013436 (rank : 107) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99689, O00679, O00728, Q6IBI7 | Gene names | FEZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.013313 (rank : 108) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.012114 (rank : 109) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.011983 (rank : 110) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ADA18_MOUSE
|
||||||
| NC score | 0.011644 (rank : 111) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R157, Q60621 | Gene names | Adam18, Adam27, Dtgn3, Tmdc3 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 18 precursor (A disintegrin and metalloproteinase domain 18) (Transmembrane metalloproteinase-like, disintegrin-like, and cysteine- rich protein III) (tMDC III) (ADAM 27). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.011608 (rank : 112) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.010956 (rank : 113) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.010912 (rank : 114) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.010764 (rank : 115) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.010385 (rank : 116) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
RAG2_MOUSE
|
||||||
| NC score | 0.009519 (rank : 117) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
TRI27_HUMAN
|
||||||
| NC score | 0.009516 (rank : 118) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14373, Q5ST26, Q6LA73, Q6NXR9, Q9BZY6, Q9UJL3 | Gene names | TRIM27, RFP, RNF76 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein RFP (Ret finger protein) (Tripartite motif- containing protein 27) (RING finger protein 76). | |||||
|
TRI27_MOUSE
|
||||||
| NC score | 0.008877 (rank : 119) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62158, Q62157, Q8C2Q5, Q99LK1 | Gene names | Trim27, Rfp | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein RFP (Ret finger protein) (Tripartite motif- containing protein 27). | |||||
|
RO52_HUMAN
|
||||||
| NC score | 0.007446 (rank : 120) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19474, Q96RF8 | Gene names | TRIM21, RNF81, RO52, SSA1 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa Ro protein (Sjoegren syndrome type A antigen) (SS-A) (Ro(SS-A)) (52 kDa ribonucleoprotein autoantigen Ro/SS-A) (Tripartite motif- containing protein 21) (RING finger protein 81). | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.007216 (rank : 121) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
DMXL1_HUMAN
|
||||||
| NC score | 0.006225 (rank : 122) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
PLXA4_HUMAN
|
||||||
| NC score | 0.006055 (rank : 123) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
FBXL7_HUMAN
|
||||||
| NC score | 0.005282 (rank : 124) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJT9, O94926 | Gene names | FBXL7, FBL6, FBL7, KIAA0840 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 7 (F-box and leucine-rich repeat protein 7) (F-box protein FBL6/FBL7). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.004659 (rank : 125) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.003434 (rank : 126) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.002998 (rank : 127) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
GRID1_HUMAN
|
||||||
| NC score | 0.002656 (rank : 128) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.002609 (rank : 129) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.002601 (rank : 130) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.000553 (rank : 131) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | -0.001355 (rank : 132) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | -0.001527 (rank : 133) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
FLT3_MOUSE
|
||||||
| NC score | -0.001891 (rank : 134) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00342 | Gene names | Flt3, Flk-2, Flt-3 | |||
|
Domain Architecture |

|
|||||
| Description | FL cytokine receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor flk-2) (Fetal liver kinase 2) (Tyrosine-protein kinase FLT3) (CD135 antigen). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | -0.003179 (rank : 135) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||