Please be patient as the page loads
|
CUL3_HUMAN
|
||||||
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CUL3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999748 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL4B_HUMAN
|
||||||
| θ value | 1.86219e-138 (rank : 3) | NC score | 0.956849 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CUL4A_MOUSE
|
||||||
| θ value | 2.4321e-138 (rank : 4) | NC score | 0.960081 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4A_HUMAN
|
||||||
| θ value | 3.51195e-137 (rank : 5) | NC score | 0.958609 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |
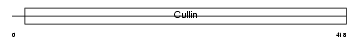
|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL1_HUMAN
|
||||||
| θ value | 9.035e-101 (rank : 6) | NC score | 0.937560 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| θ value | 9.035e-101 (rank : 7) | NC score | 0.937553 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL2_HUMAN
|
||||||
| θ value | 8.20983e-86 (rank : 8) | NC score | 0.931961 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13617, O00200, Q9UNF9 | Gene names | CUL2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL2_MOUSE
|
||||||
| θ value | 3.56943e-81 (rank : 9) | NC score | 0.930688 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D4H8, Q3TUR8 | Gene names | Cul2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL5_HUMAN
|
||||||
| θ value | 1.96772e-63 (rank : 10) | NC score | 0.904227 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q93034, O14766, Q9BZC6 | Gene names | CUL5, VACM1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5) (Vasopressin-activated calcium-mobilizing receptor) (VACM-1). | |||||
|
CUL5_MOUSE
|
||||||
| θ value | 2.84138e-62 (rank : 11) | NC score | 0.904197 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D5V5, Q8BMQ6, Q8BV53, Q8C098 | Gene names | Cul5 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5). | |||||
|
CJ046_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 12) | NC score | 0.200469 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86Y37, Q5XPL7, Q8IY11, Q8N7S4 | Gene names | C10orf46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46. | |||||
|
CJ046_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 13) | NC score | 0.201555 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0X2, Q3TE79, Q9CY95 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46 homolog. | |||||
|
ANC2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 14) | NC score | 0.387226 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZQ7, Q8R2Q1 | Gene names | Anapc2 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
ANC2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 15) | NC score | 0.383927 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJX6, Q5VSG1, Q96DG5, Q96GG4, Q9P2E1 | Gene names | ANAPC2, APC2, KIAA1406 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.084673 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
A4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.032957 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.016662 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
PFTA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.041096 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49354, Q9UDC1 | Gene names | FNTA | |||
|
Domain Architecture |
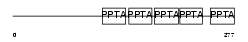
|
|||||
| Description | Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit (EC 2.5.1.58) (EC 2.5.1.59) (CAAX farnesyltransferase alpha subunit) (Ras proteins prenyltransferase alpha) (FTase-alpha) (Type I protein geranyl-geranyltransferase alpha subunit) (GGTase-I-alpha). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.012526 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CTGE2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.019655 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
A4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.027094 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.010004 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.007967 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.011804 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.008034 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
NOL8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.019318 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.008566 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.016465 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.015486 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.005634 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.007097 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.006371 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CM007_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.012700 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
NPHP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.013941 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75161 | Gene names | NPHP4, KIAA0673 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.009643 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
CLUA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.014017 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
COE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.004487 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08792, Q543D5 | Gene names | Ebf2, Coe2, Mmot1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE2 (Early B-cell factor 2) (EBF-2) (Olf-1/EBF- like 3) (OE-3) (O/E-3) (Metencephalon-mesencephalon-olfactory transcription factor 1) (MET-mesencephalon-olfactory transcription factor 1) (MET-mesencephalon-olfactory TF1). | |||||
|
DCP1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.008389 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZD4, Q86XH9, Q96BP8, Q96MZ8 | Gene names | DCP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1B (EC 3.-.-.-). | |||||
|
DDX58_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.010721 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.014238 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
SYCC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.007661 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49589, Q53XI8, Q9HD24, Q9HD25 | Gene names | CARS | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
CUL7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.063572 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
PARC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.055196 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
CUL3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_MOUSE
|
||||||
| NC score | 0.999748 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL4A_MOUSE
|
||||||
| NC score | 0.960081 (rank : 3) | θ value | 2.4321e-138 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4A_HUMAN
|
||||||
| NC score | 0.958609 (rank : 4) | θ value | 3.51195e-137 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4B_HUMAN
|
||||||
| NC score | 0.956849 (rank : 5) | θ value | 1.86219e-138 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CUL1_HUMAN
|
||||||
| NC score | 0.937560 (rank : 6) | θ value | 9.035e-101 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| NC score | 0.937553 (rank : 7) | θ value | 9.035e-101 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL2_HUMAN
|
||||||
| NC score | 0.931961 (rank : 8) | θ value | 8.20983e-86 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13617, O00200, Q9UNF9 | Gene names | CUL2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL2_MOUSE
|
||||||
| NC score | 0.930688 (rank : 9) | θ value | 3.56943e-81 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D4H8, Q3TUR8 | Gene names | Cul2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL5_HUMAN
|
||||||
| NC score | 0.904227 (rank : 10) | θ value | 1.96772e-63 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q93034, O14766, Q9BZC6 | Gene names | CUL5, VACM1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5) (Vasopressin-activated calcium-mobilizing receptor) (VACM-1). | |||||
|
CUL5_MOUSE
|
||||||
| NC score | 0.904197 (rank : 11) | θ value | 2.84138e-62 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D5V5, Q8BMQ6, Q8BV53, Q8C098 | Gene names | Cul5 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5). | |||||
|
ANC2_MOUSE
|
||||||
| NC score | 0.387226 (rank : 12) | θ value | 0.000461057 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZQ7, Q8R2Q1 | Gene names | Anapc2 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
ANC2_HUMAN
|
||||||
| NC score | 0.383927 (rank : 13) | θ value | 0.00134147 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJX6, Q5VSG1, Q96DG5, Q96GG4, Q9P2E1 | Gene names | ANAPC2, APC2, KIAA1406 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
CJ046_MOUSE
|
||||||
| NC score | 0.201555 (rank : 14) | θ value | 2.88788e-06 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0X2, Q3TE79, Q9CY95 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46 homolog. | |||||
|
CJ046_HUMAN
|
||||||
| NC score | 0.200469 (rank : 15) | θ value | 2.88788e-06 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86Y37, Q5XPL7, Q8IY11, Q8N7S4 | Gene names | C10orf46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46. | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.084673 (rank : 16) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.063572 (rank : 17) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
PARC_MOUSE
|
||||||
| NC score | 0.055196 (rank : 18) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
PFTA_HUMAN
|
||||||
| NC score | 0.041096 (rank : 19) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49354, Q9UDC1 | Gene names | FNTA | |||
|
Domain Architecture |

|
|||||
| Description | Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit (EC 2.5.1.58) (EC 2.5.1.59) (CAAX farnesyltransferase alpha subunit) (Ras proteins prenyltransferase alpha) (FTase-alpha) (Type I protein geranyl-geranyltransferase alpha subunit) (GGTase-I-alpha). | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.032957 (rank : 20) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.027094 (rank : 21) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
CTGE2_HUMAN
|
||||||
| NC score | 0.019655 (rank : 22) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
NOL8_MOUSE
|
||||||
| NC score | 0.019318 (rank : 23) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.016662 (rank : 24) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.016465 (rank : 25) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.015486 (rank : 26) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.014238 (rank : 27) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
CLUA1_MOUSE
|
||||||
| NC score | 0.014017 (rank : 28) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
NPHP4_HUMAN
|
||||||
| NC score | 0.013941 (rank : 29) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75161 | Gene names | NPHP4, KIAA0673 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.012700 (rank : 30) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.012526 (rank : 31) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.011804 (rank : 32) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.010721 (rank : 33) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.010004 (rank : 34) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.009643 (rank : 35) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.008566 (rank : 36) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
DCP1B_HUMAN
|
||||||
| NC score | 0.008389 (rank : 37) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZD4, Q86XH9, Q96BP8, Q96MZ8 | Gene names | DCP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1B (EC 3.-.-.-). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.008034 (rank : 38) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.007967 (rank : 39) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
SYCC_HUMAN
|
||||||
| NC score | 0.007661 (rank : 40) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49589, Q53XI8, Q9HD24, Q9HD25 | Gene names | CARS | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.007097 (rank : 41) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.006371 (rank : 42) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.005634 (rank : 43) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
COE2_MOUSE
|
||||||
| NC score | 0.004487 (rank : 44) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08792, Q543D5 | Gene names | Ebf2, Coe2, Mmot1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE2 (Early B-cell factor 2) (EBF-2) (Olf-1/EBF- like 3) (OE-3) (O/E-3) (Metencephalon-mesencephalon-olfactory transcription factor 1) (MET-mesencephalon-olfactory transcription factor 1) (MET-mesencephalon-olfactory TF1). | |||||