Please be patient as the page loads
|
ANC2_MOUSE
|
||||||
| SwissProt Accessions | Q8BZQ7, Q8R2Q1 | Gene names | Anapc2 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ANC2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998369 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UJX6, Q5VSG1, Q96DG5, Q96GG4, Q9P2E1 | Gene names | ANAPC2, APC2, KIAA1406 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
ANC2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BZQ7, Q8R2Q1 | Gene names | Anapc2 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
CUL4A_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 3) | NC score | 0.457123 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |
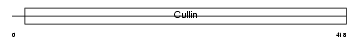
|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4A_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 4) | NC score | 0.457465 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4B_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 5) | NC score | 0.451459 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CUL1_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 6) | NC score | 0.432064 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 7) | NC score | 0.432050 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL2_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 8) | NC score | 0.415233 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13617, O00200, Q9UNF9 | Gene names | CUL2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL2_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 9) | NC score | 0.415225 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D4H8, Q3TUR8 | Gene names | Cul2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 10) | NC score | 0.387226 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 11) | NC score | 0.387266 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL5_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 12) | NC score | 0.363652 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q93034, O14766, Q9BZC6 | Gene names | CUL5, VACM1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5) (Vasopressin-activated calcium-mobilizing receptor) (VACM-1). | |||||
|
CUL5_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.360526 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D5V5, Q8BMQ6, Q8BV53, Q8C098 | Gene names | Cul5 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.031850 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.028223 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
GNA15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.019293 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30679, O75247 | Gene names | GNA15, GNA16 | |||
|
Domain Architecture |
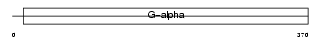
|
|||||
| Description | Guanine nucleotide-binding protein alpha-15 subunit (G alpha-15) (G alpha-16). | |||||
|
ITA10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.017139 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75578, Q6UXJ6, Q9UHZ8 | Gene names | ITGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-10 precursor. | |||||
|
PROM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.040028 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43490 | Gene names | PROM1, PROML1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133) (CD133 antigen). | |||||
|
HEAT3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.035482 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |
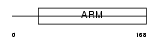
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
PPWD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.014595 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CEC6 | Gene names | Ppwd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.009862 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
CEBPZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.012747 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03701, Q8NE75 | Gene names | CEBPZ, CBF2 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
GAK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.003417 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.009364 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
STAT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.006225 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
ANC2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BZQ7, Q8R2Q1 | Gene names | Anapc2 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
ANC2_HUMAN
|
||||||
| NC score | 0.998369 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UJX6, Q5VSG1, Q96DG5, Q96GG4, Q9P2E1 | Gene names | ANAPC2, APC2, KIAA1406 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
CUL4A_MOUSE
|
||||||
| NC score | 0.457465 (rank : 3) | θ value | 3.29651e-10 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4A_HUMAN
|
||||||
| NC score | 0.457123 (rank : 4) | θ value | 1.9326e-10 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4B_HUMAN
|
||||||
| NC score | 0.451459 (rank : 5) | θ value | 4.30538e-10 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CUL1_HUMAN
|
||||||
| NC score | 0.432064 (rank : 6) | θ value | 6.87365e-08 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| NC score | 0.432050 (rank : 7) | θ value | 6.87365e-08 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL2_HUMAN
|
||||||
| NC score | 0.415233 (rank : 8) | θ value | 2.21117e-06 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13617, O00200, Q9UNF9 | Gene names | CUL2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL2_MOUSE
|
||||||
| NC score | 0.415225 (rank : 9) | θ value | 2.88788e-06 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D4H8, Q3TUR8 | Gene names | Cul2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL3_MOUSE
|
||||||
| NC score | 0.387266 (rank : 10) | θ value | 0.000461057 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_HUMAN
|
||||||
| NC score | 0.387226 (rank : 11) | θ value | 0.000461057 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL5_HUMAN
|
||||||
| NC score | 0.363652 (rank : 12) | θ value | 0.00390308 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q93034, O14766, Q9BZC6 | Gene names | CUL5, VACM1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5) (Vasopressin-activated calcium-mobilizing receptor) (VACM-1). | |||||
|
CUL5_MOUSE
|
||||||
| NC score | 0.360526 (rank : 13) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D5V5, Q8BMQ6, Q8BV53, Q8C098 | Gene names | Cul5 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5). | |||||
|
PROM1_HUMAN
|
||||||
| NC score | 0.040028 (rank : 14) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43490 | Gene names | PROM1, PROML1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133) (CD133 antigen). | |||||
|
HEAT3_MOUSE
|
||||||
| NC score | 0.035482 (rank : 15) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.031850 (rank : 16) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.028223 (rank : 17) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
GNA15_HUMAN
|
||||||
| NC score | 0.019293 (rank : 18) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30679, O75247 | Gene names | GNA15, GNA16 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein alpha-15 subunit (G alpha-15) (G alpha-16). | |||||
|
ITA10_HUMAN
|
||||||
| NC score | 0.017139 (rank : 19) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75578, Q6UXJ6, Q9UHZ8 | Gene names | ITGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-10 precursor. | |||||
|
PPWD1_MOUSE
|
||||||
| NC score | 0.014595 (rank : 20) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CEC6 | Gene names | Ppwd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidylprolyl isomerase domain and WD repeat-containing protein 1 (EC 5.2.1.8). | |||||
|
CEBPZ_HUMAN
|
||||||
| NC score | 0.012747 (rank : 21) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03701, Q8NE75 | Gene names | CEBPZ, CBF2 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.009862 (rank : 22) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.009364 (rank : 23) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
STAT1_HUMAN
|
||||||
| NC score | 0.006225 (rank : 24) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.003417 (rank : 25) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||