Please be patient as the page loads
|
DDX58_HUMAN
|
||||||
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DDX58_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984004 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 5.29683e-101 (rank : 3) | NC score | 0.895503 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 8.75112e-96 (rank : 4) | NC score | 0.893349 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | 4.07447e-85 (rank : 5) | NC score | 0.890290 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 1.35638e-80 (rank : 6) | NC score | 0.893766 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
DICER_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 7) | NC score | 0.587370 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 8) | NC score | 0.587644 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 9) | NC score | 0.539320 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 10) | NC score | 0.555134 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
HELC1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 11) | NC score | 0.240831 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.218458 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 13) | NC score | 0.217884 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.201476 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 15) | NC score | 0.212560 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 16) | NC score | 0.212587 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 17) | NC score | 0.212946 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 18) | NC score | 0.212946 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 19) | NC score | 0.215542 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 20) | NC score | 0.215542 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
U520_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 21) | NC score | 0.195034 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 22) | NC score | 0.030165 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.182712 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.208556 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 25) | NC score | 0.208740 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.198404 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.017738 (rank : 116) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.028898 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.032487 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.204846 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 31) | NC score | 0.202304 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.147385 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.197706 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.206004 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.206010 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.031732 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.031290 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.163037 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.192333 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.176704 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.200618 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.198415 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX56_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.147890 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
PEX5R_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.031509 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.201666 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.199448 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.193650 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DFFA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.033563 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54786, O54787 | Gene names | Dffa, Icad | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA fragmentation factor subunit alpha (DNA fragmentation factor 45 kDa subunit) (DFF-45) (Inhibitor of CAD) (ICAD). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.015278 (rank : 122) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.201129 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
PRLPI_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.024884 (rank : 107) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUN5, Q5SZY4 | Gene names | Prlpi, Prlpj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental prolactin-like protein I precursor (PRL-like protein I) (PLP-I) (Decidualin) (Prolactin-like protein J) (PRL-like protein J) (PLP-J). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.017570 (rank : 117) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.194360 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.197067 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.159343 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.018494 (rank : 111) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
POLH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.032606 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJN0, Q9JJJ2 | Gene names | Polh, Rad30a, Xpv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein homolog). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.192537 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.158761 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
PEX5R_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.027629 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.031531 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CUL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.013416 (rank : 125) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.186251 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.202711 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.197739 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
ORC6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.023080 (rank : 108) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUJ8 | Gene names | Orc6l, Orc6 | |||
|
Domain Architecture |
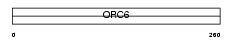
|
|||||
| Description | Origin recognition complex subunit 6. | |||||
|
POLI_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.028664 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNA4, Q8N590, Q9H0S1, Q9NYH6 | Gene names | POLI, RAD30B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (RAD30 homolog B) (Eta2). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.011567 (rank : 129) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.018594 (rank : 110) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.188008 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.188440 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.017892 (rank : 115) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
POLI_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.027504 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6R3M4, Q641P1, Q6R3M3, Q9R1A6 | Gene names | Poli, Rad30b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (Rad30 homolog B). | |||||
|
BTK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | -0.002250 (rank : 136) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06187 | Gene names | BTK, AGMX1, ATK, BPK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase BTK (EC 2.7.10.2) (Bruton tyrosine kinase) (Agammaglobulinaemia tyrosine kinase) (ATK) (B cell progenitor kinase) (BPK). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.198096 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.015013 (rank : 123) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.005637 (rank : 135) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CCD91_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.015973 (rank : 120) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.180372 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.172212 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.169597 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.016378 (rank : 119) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.196064 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
POLH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.026158 (rank : 106) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y253, Q7L8E3, Q96BC4, Q9BX13 | Gene names | POLH, RAD30, RAD30A, XPV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.005674 (rank : 133) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.005834 (rank : 132) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.017968 (rank : 114) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
AQR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.029936 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
AQR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.035421 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.027821 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.017152 (rank : 118) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CUL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.010721 (rank : 130) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.161183 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.160717 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
K6PP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.005662 (rank : 134) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUA3, Q3TNA9, Q3U4P1, Q3U7G4, Q4KUG1, Q543K8, Q8C5I6, Q9JI86 | Gene names | Pfkp, Pfkc | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructokinase type C (EC 2.7.1.11) (Phosphofructokinase 1) (Phosphohexokinase) (Phosphofructo-1-kinase isozyme C) (PFK-C). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.014966 (rank : 124) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.006943 (rank : 131) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
RNF8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.011854 (rank : 128) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O76064 | Gene names | RNF8, KIAA0646 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.013362 (rank : 127) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.013395 (rank : 126) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.018415 (rank : 112) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
STIM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.018723 (rank : 109) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
STIM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.018186 (rank : 113) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TF3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | -0.002864 (rank : 137) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92664, Q12963, Q13097 | Gene names | GTF3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor IIIA (Factor A) (TFIIIA). | |||||
|
TNNT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.015597 (rank : 121) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
BLM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.067117 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
BLM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.067841 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.161726 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.161968 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.161397 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.146078 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.144580 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.149264 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.154205 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.155215 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.145058 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.144430 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.153440 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.151860 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.175728 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.178230 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.193102 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.186084 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.142802 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.159483 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.163937 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.160709 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
RECQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.084626 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.079563 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.142028 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.130287 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.086943 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
RNC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.056193 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
UAP56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.155168 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.155098 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
WRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.075358 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
WRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.079847 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.984004 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.895503 (rank : 3) | θ value | 5.29683e-101 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.893766 (rank : 4) | θ value | 1.35638e-80 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.893349 (rank : 5) | θ value | 8.75112e-96 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.890290 (rank : 6) | θ value | 4.07447e-85 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.587644 (rank : 7) | θ value | 1.37858e-24 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.587370 (rank : 8) | θ value | 4.73814e-25 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.555134 (rank : 9) | θ value | 1.74391e-19 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.539320 (rank : 10) | θ value | 1.02238e-19 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.240831 (rank : 11) | θ value | 9.29e-05 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.218458 (rank : 12) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.217884 (rank : 13) | θ value | 0.000602161 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.215542 (rank : 14) | θ value | 0.0113563 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.215542 (rank : 15) | θ value | 0.0113563 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.212946 (rank : 16) | θ value | 0.00869519 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.212946 (rank : 17) | θ value | 0.00869519 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.212587 (rank : 18) | θ value | 0.00665767 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.212560 (rank : 19) | θ value | 0.00665767 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.208740 (rank : 20) | θ value | 0.0330416 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.208556 (rank : 21) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.206010 (rank : 22) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.206004 (rank : 23) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.204846 (rank : 24) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.202711 (rank : 25) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.202304 (rank : 26) | θ value | 0.125558 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.201666 (rank : 27) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.201476 (rank : 28) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.201129 (rank : 29) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.200618 (rank : 30) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.199448 (rank : 31) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.198415 (rank : 32) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.198404 (rank : 33) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.198096 (rank : 34) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.197739 (rank : 35) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.197706 (rank : 36) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.197067 (rank : 37) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.196064 (rank : 38) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.195034 (rank : 39) | θ value | 0.0113563 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.194360 (rank : 40) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.193650 (rank : 41) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.193102 (rank : 42) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.192537 (rank : 43) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.192333 (rank : 44) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.188440 (rank : 45) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.188008 (rank : 46) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.186251 (rank : 47) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.186084 (rank : 48) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.182712 (rank : 49) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.180372 (rank : 50) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.178230 (rank : 51) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.176704 (rank : 52) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.175728 (rank : 53) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.172212 (rank : 54) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.169597 (rank : 55) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.163937 (rank : 56) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.163037 (rank : 57) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.161968 (rank : 58) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.161726 (rank : 59) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.161397 (rank : 60) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.161183 (rank : 61) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.160717 (rank : 62) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.160709 (rank : 63) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.159483 (rank : 64) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.159343 (rank : 65) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.158761 (rank : 66) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.155215 (rank : 67) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.155168 (rank : 68) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.155098 (rank : 69) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.154205 (rank : 70) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.153440 (rank : 71) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.151860 (rank : 72) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.149264 (rank : 73) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.147890 (rank : 74) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.147385 (rank : 75) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.146078 (rank : 76) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.145058 (rank : 77) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.144580 (rank : 78) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.144430 (rank : 79) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.142802 (rank : 80) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.142028 (rank : 81) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.130287 (rank : 82) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.086943 (rank : 83) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.084626 (rank : 84) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.079847 (rank : 85) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.079563 (rank : 86) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.075358 (rank : 87) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.067841 (rank : 88) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.067117 (rank : 89) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.056193 (rank : 90) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
AQR_MOUSE
|
||||||
| NC score | 0.035421 (rank : 91) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
DFFA_MOUSE
|
||||||
| NC score | 0.033563 (rank : 92) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54786, O54787 | Gene names | Dffa, Icad | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA fragmentation factor subunit alpha (DNA fragmentation factor 45 kDa subunit) (DFF-45) (Inhibitor of CAD) (ICAD). | |||||
|
POLH_MOUSE
|
||||||
| NC score | 0.032606 (rank : 93) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJN0, Q9JJJ2 | Gene names | Polh, Rad30a, Xpv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein homolog). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.032487 (rank : 94) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.031732 (rank : 95) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.031531 (rank : 96) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
PEX5R_HUMAN
|
||||||
| NC score | 0.031509 (rank : 97) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.031290 (rank : 98) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.030165 (rank : 99) | θ value | 0.0252991 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
AQR_HUMAN
|
||||||
| NC score | 0.029936 (rank : 100) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.028898 (rank : 101) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
POLI_HUMAN
|
||||||
| NC score | 0.028664 (rank : 102) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNA4, Q8N590, Q9H0S1, Q9NYH6 | Gene names | POLI, RAD30B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (RAD30 homolog B) (Eta2). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.027821 (rank : 103) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
PEX5R_MOUSE
|
||||||
| NC score | 0.027629 (rank : 104) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
POLI_MOUSE
|
||||||
| NC score | 0.027504 (rank : 105) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6R3M4, Q641P1, Q6R3M3, Q9R1A6 | Gene names | Poli, Rad30b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (Rad30 homolog B). | |||||
|
POLH_HUMAN
|
||||||
| NC score | 0.026158 (rank : 106) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y253, Q7L8E3, Q96BC4, Q9BX13 | Gene names | POLH, RAD30, RAD30A, XPV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein). | |||||
|
PRLPI_MOUSE
|
||||||
| NC score | 0.024884 (rank : 107) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUN5, Q5SZY4 | Gene names | Prlpi, Prlpj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental prolactin-like protein I precursor (PRL-like protein I) (PLP-I) (Decidualin) (Prolactin-like protein J) (PRL-like protein J) (PLP-J). | |||||
|
ORC6_MOUSE
|
||||||
| NC score | 0.023080 (rank : 108) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUJ8 | Gene names | Orc6l, Orc6 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 6. | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.018723 (rank : 109) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.018594 (rank : 110) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.018494 (rank : 111) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.018415 (rank : 112) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
STIM1_MOUSE
|
||||||
| NC score | 0.018186 (rank : 113) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.017968 (rank : 114) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.017892 (rank : 115) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.017738 (rank : 116) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.017570 (rank : 117) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.017152 (rank : 118) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.016378 (rank : 119) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
CCD91_MOUSE
|
||||||
| NC score | 0.015973 (rank : 120) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
TNNT2_HUMAN
|
||||||
| NC score | 0.015597 (rank : 121) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.015278 (rank : 122) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.015013 (rank : 123) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.014966 (rank : 124) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CUL3_MOUSE
|
||||||
| NC score | 0.013416 (rank : 125) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.013395 (rank : 126) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.013362 (rank : 127) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
RNF8_HUMAN
|
||||||
| NC score | 0.011854 (rank : 128) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O76064 | Gene names | RNF8, KIAA0646 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.011567 (rank : 129) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
CUL3_HUMAN
|
||||||
| NC score | 0.010721 (rank : 130) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.006943 (rank : 131) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.005834 (rank : 132) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.005674 (rank : 133) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
K6PP_MOUSE
|
||||||
| NC score | 0.005662 (rank : 134) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUA3, Q3TNA9, Q3U4P1, Q3U7G4, Q4KUG1, Q543K8, Q8C5I6, Q9JI86 | Gene names | Pfkp, Pfkc | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructokinase type C (EC 2.7.1.11) (Phosphofructokinase 1) (Phosphohexokinase) (Phosphofructo-1-kinase isozyme C) (PFK-C). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.005637 (rank : 135) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BTK_HUMAN
|
||||||
| NC score | -0.002250 (rank : 136) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06187 | Gene names | BTK, AGMX1, ATK, BPK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase BTK (EC 2.7.10.2) (Bruton tyrosine kinase) (Agammaglobulinaemia tyrosine kinase) (ATK) (B cell progenitor kinase) (BPK). | |||||
|
TF3A_HUMAN
|
||||||
| NC score | -0.002864 (rank : 137) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92664, Q12963, Q13097 | Gene names | GTF3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor IIIA (Factor A) (TFIIIA). | |||||