Please be patient as the page loads
|
DDX54_HUMAN
|
||||||
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DDX54_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 170 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995115 (rank : 2) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 1.45929e-58 (rank : 3) | NC score | 0.967349 (rank : 4) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 2.10721e-57 (rank : 4) | NC score | 0.964015 (rank : 6) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 2.32979e-56 (rank : 5) | NC score | 0.942543 (rank : 49) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 2.32979e-56 (rank : 6) | NC score | 0.940439 (rank : 50) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 5.19021e-56 (rank : 7) | NC score | 0.961595 (rank : 7) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 4.39379e-55 (rank : 8) | NC score | 0.960058 (rank : 10) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 4.85792e-54 (rank : 9) | NC score | 0.967147 (rank : 5) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 4.85792e-54 (rank : 10) | NC score | 0.967656 (rank : 3) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 7.01481e-53 (rank : 11) | NC score | 0.932224 (rank : 54) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 5.55816e-50 (rank : 12) | NC score | 0.954551 (rank : 17) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 1.61718e-49 (rank : 13) | NC score | 0.953320 (rank : 20) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 2.1121e-49 (rank : 14) | NC score | 0.953153 (rank : 21) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 4.70527e-49 (rank : 15) | NC score | 0.961150 (rank : 8) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 4.70527e-49 (rank : 16) | NC score | 0.960976 (rank : 9) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 1.788e-48 (rank : 17) | NC score | 0.954027 (rank : 18) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 1.51363e-47 (rank : 18) | NC score | 0.954895 (rank : 14) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 1.97686e-47 (rank : 19) | NC score | 0.947405 (rank : 38) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 2.58187e-47 (rank : 20) | NC score | 0.947342 (rank : 40) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 2.58187e-47 (rank : 21) | NC score | 0.944661 (rank : 46) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 4.40402e-47 (rank : 22) | NC score | 0.944322 (rank : 47) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 7.51208e-47 (rank : 23) | NC score | 0.944925 (rank : 45) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 24) | NC score | 0.945112 (rank : 44) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 1.28137e-46 (rank : 25) | NC score | 0.956833 (rank : 12) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 6.35935e-46 (rank : 26) | NC score | 0.949923 (rank : 28) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 8.30557e-46 (rank : 27) | NC score | 0.952735 (rank : 23) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 8.30557e-46 (rank : 28) | NC score | 0.952870 (rank : 22) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 3.15612e-45 (rank : 29) | NC score | 0.956946 (rank : 11) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 3.15612e-45 (rank : 30) | NC score | 0.956460 (rank : 13) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 3.15612e-45 (rank : 31) | NC score | 0.949885 (rank : 29) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 4.122e-45 (rank : 32) | NC score | 0.942816 (rank : 48) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 2.04573e-44 (rank : 33) | NC score | 0.953513 (rank : 19) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 4.55743e-44 (rank : 34) | NC score | 0.949672 (rank : 30) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 4.55743e-44 (rank : 35) | NC score | 0.949672 (rank : 31) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 2.26182e-43 (rank : 36) | NC score | 0.949125 (rank : 33) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 2.26182e-43 (rank : 37) | NC score | 0.949125 (rank : 34) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | 1.12253e-42 (rank : 38) | NC score | 0.952174 (rank : 24) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 5.21438e-40 (rank : 39) | NC score | 0.954803 (rank : 16) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 6.81017e-40 (rank : 40) | NC score | 0.948321 (rank : 36) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 6.81017e-40 (rank : 41) | NC score | 0.954827 (rank : 15) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 5.76516e-39 (rank : 42) | NC score | 0.952011 (rank : 25) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
UAP56_HUMAN
|
||||||
| θ value | 5.76516e-39 (rank : 43) | NC score | 0.947082 (rank : 41) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | 5.76516e-39 (rank : 44) | NC score | 0.947002 (rank : 43) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 45) | NC score | 0.951200 (rank : 26) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | 3.73686e-38 (rank : 46) | NC score | 0.947348 (rank : 39) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | 1.85459e-37 (rank : 47) | NC score | 0.947506 (rank : 37) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 7.04741e-37 (rank : 48) | NC score | 0.926269 (rank : 61) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 7.04741e-37 (rank : 49) | NC score | 0.938612 (rank : 51) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 1.57e-36 (rank : 50) | NC score | 0.938514 (rank : 52) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 1.01765e-35 (rank : 51) | NC score | 0.918506 (rank : 65) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 3.86705e-35 (rank : 52) | NC score | 0.951166 (rank : 27) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | 2.50655e-34 (rank : 53) | NC score | 0.949011 (rank : 35) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX56_HUMAN
|
||||||
| θ value | 1.6247e-33 (rank : 54) | NC score | 0.949258 (rank : 32) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 55) | NC score | 0.928588 (rank : 58) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 1.79631e-32 (rank : 56) | NC score | 0.935633 (rank : 53) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 2.34606e-32 (rank : 57) | NC score | 0.932194 (rank : 55) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 3.06405e-32 (rank : 58) | NC score | 0.927428 (rank : 60) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 59) | NC score | 0.926148 (rank : 62) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 60) | NC score | 0.925540 (rank : 63) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 1.16434e-31 (rank : 61) | NC score | 0.925442 (rank : 64) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 62) | NC score | 0.930346 (rank : 56) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 63) | NC score | 0.927540 (rank : 59) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 1.08979e-29 (rank : 64) | NC score | 0.928695 (rank : 57) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 1.4233e-29 (rank : 65) | NC score | 0.947031 (rank : 42) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 66) | NC score | 0.911196 (rank : 66) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 3.17815e-21 (rank : 67) | NC score | 0.884089 (rank : 67) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 68) | NC score | 0.882427 (rank : 68) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
WRN_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 69) | NC score | 0.470921 (rank : 73) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
BLM_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 70) | NC score | 0.492944 (rank : 71) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 71) | NC score | 0.040703 (rank : 100) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
BLM_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 72) | NC score | 0.479116 (rank : 72) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 73) | NC score | 0.435904 (rank : 75) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 74) | NC score | 0.024765 (rank : 114) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 75) | NC score | 0.024023 (rank : 116) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 76) | NC score | 0.052233 (rank : 98) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 77) | NC score | 0.517358 (rank : 70) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 78) | NC score | 0.041084 (rank : 99) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
SK2L2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 79) | NC score | 0.066263 (rank : 94) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42285, Q8TAG2 | Gene names | SKIV2L2, KIAA0052 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
SK2L2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 80) | NC score | 0.066072 (rank : 95) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CZU3 | Gene names | Skiv2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
SKIV2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 81) | NC score | 0.087577 (rank : 92) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
PRP16_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 82) | NC score | 0.027397 (rank : 110) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 83) | NC score | 0.530729 (rank : 69) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
CIR_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 84) | NC score | 0.040566 (rank : 101) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 85) | NC score | 0.025434 (rank : 112) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
RECQ1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 86) | NC score | 0.423293 (rank : 76) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 87) | NC score | 0.024623 (rank : 115) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 88) | NC score | 0.450674 (rank : 74) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 89) | NC score | 0.008518 (rank : 135) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 90) | NC score | 0.313198 (rank : 82) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
T2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 91) | NC score | 0.040298 (rank : 102) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
HAP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 92) | NC score | 0.013412 (rank : 129) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35668, O35636 | Gene names | Hap1 | |||
|
Domain Architecture |
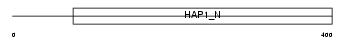
|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 93) | NC score | 0.420316 (rank : 77) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 94) | NC score | 0.025303 (rank : 113) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 95) | NC score | 0.297822 (rank : 85) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
CT004_MOUSE
|
||||||
| θ value | 0.62314 (rank : 96) | NC score | 0.017290 (rank : 121) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D2V5, Q3TM16, Q8VBU9 | Gene names | ||||
|
Domain Architecture |
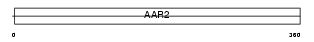
|
|||||
| Description | Protein C20orf4 homolog. | |||||
|
EI2BD_MOUSE
|
||||||
| θ value | 0.62314 (rank : 97) | NC score | 0.014826 (rank : 126) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61749, Q61748, Q8VC35 | Gene names | Eif2b4, Eif2b, Eif2bd, Jgr1a | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 0.62314 (rank : 98) | NC score | 0.006707 (rank : 144) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
RU17_HUMAN
|
||||||
| θ value | 0.62314 (rank : 99) | NC score | 0.017443 (rank : 120) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.62314 (rank : 100) | NC score | 0.004275 (rank : 160) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 101) | NC score | 0.003729 (rank : 166) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 0.813845 (rank : 102) | NC score | 0.008504 (rank : 136) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
DDX58_MOUSE
|
||||||
| θ value | 1.06291 (rank : 103) | NC score | 0.191322 (rank : 86) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
CC026_MOUSE
|
||||||
| θ value | 1.38821 (rank : 104) | NC score | 0.133451 (rank : 90) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 105) | NC score | 0.014470 (rank : 127) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
H2BWT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 106) | NC score | 0.006283 (rank : 150) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2G1 | Gene names | H2BFWT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2B type W-T (H2B histone family member W testis-specific). | |||||
|
WDR60_MOUSE
|
||||||
| θ value | 1.38821 (rank : 107) | NC score | 0.012718 (rank : 130) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
DDX58_HUMAN
|
||||||
| θ value | 1.81305 (rank : 108) | NC score | 0.159343 (rank : 88) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DICER_HUMAN
|
||||||
| θ value | 1.81305 (rank : 109) | NC score | 0.373506 (rank : 80) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 110) | NC score | -0.004901 (rank : 175) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
RN169_HUMAN
|
||||||
| θ value | 1.81305 (rank : 111) | NC score | 0.013523 (rank : 128) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
SNW1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 112) | NC score | 0.032214 (rank : 106) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13573, Q13483, Q32N03, Q5D0D6 | Gene names | SNW1, SKIIP, SKIP | |||
|
Domain Architecture |
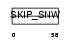
|
|||||
| Description | SNW domain-containing protein 1 (Nuclear protein SkiP) (Ski- interacting protein) (Nuclear receptor coactivator NCoA-62). | |||||
|
SNW1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 113) | NC score | 0.032284 (rank : 105) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CSN1 | Gene names | Snw1, Skiip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNW domain-containing protein 1 (Nuclear protein SkiP) (Ski- interacting protein). | |||||
|
U520_HUMAN
|
||||||
| θ value | 1.81305 (rank : 114) | NC score | 0.143968 (rank : 89) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 2.36792 (rank : 115) | NC score | 0.372853 (rank : 81) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
LC7L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 116) | NC score | 0.008962 (rank : 133) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 117) | NC score | 0.007696 (rank : 140) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
RING1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 118) | NC score | 0.004053 (rank : 162) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
RU17_MOUSE
|
||||||
| θ value | 2.36792 (rank : 119) | NC score | 0.015965 (rank : 123) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
SAPS2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 120) | NC score | 0.006147 (rank : 151) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
TR150_HUMAN
|
||||||
| θ value | 2.36792 (rank : 121) | NC score | 0.030237 (rank : 107) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 122) | NC score | 0.002911 (rank : 168) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 123) | NC score | 0.018454 (rank : 119) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
LC7L2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 124) | NC score | 0.008915 (rank : 134) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 125) | NC score | 0.299118 (rank : 84) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 126) | NC score | 0.005774 (rank : 153) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 127) | NC score | 0.001749 (rank : 170) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 128) | NC score | -0.004109 (rank : 174) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 3.0926 (rank : 129) | NC score | 0.003165 (rank : 167) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 130) | NC score | 0.003796 (rank : 165) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
2A5D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.004030 (rank : 163) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14738, O00494, O00696, Q15171 | Gene names | PPP2R5D | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (PP2A, B subunit, B' delta isoform) (PP2A, B subunit, B56 delta isoform) (PP2A, B subunit, PR61 delta isoform) (PP2A, B subunit, R5 delta isoform). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.000723 (rank : 171) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
COX10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.008499 (rank : 137) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 134) | NC score | 0.011509 (rank : 131) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
GA2L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 135) | NC score | 0.005310 (rank : 156) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 136) | NC score | 0.004077 (rank : 161) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
JIP3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 137) | NC score | 0.005942 (rank : 152) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 138) | NC score | 0.312208 (rank : 83) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
NTHL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 139) | NC score | 0.009399 (rank : 132) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78549, Q99566, Q99794, Q9BPX2 | Gene names | NTHL1, NTH1, OCTS3 | |||
|
Domain Architecture |
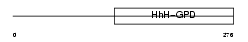
|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 140) | NC score | 0.027078 (rank : 111) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
TCAL6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 141) | NC score | 0.016340 (rank : 122) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6IPX3, Q5H9J8 | Gene names | TCEAL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 6 (TCEA-like protein 6) (Transcription elongation factor S-II protein-like 6). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 4.03905 (rank : 142) | NC score | 0.027420 (rank : 109) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.005624 (rank : 154) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.037689 (rank : 103) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.006877 (rank : 143) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
LBR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.005104 (rank : 157) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U9G9, Q3TSW2, Q811V8, Q811V9, Q8BST3, Q8K2Y8, Q8VDM0, Q91YS5, Q91Z27 | Gene names | Lbr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein). | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.007522 (rank : 141) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
PDCD7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 148) | NC score | 0.005488 (rank : 155) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTY1, Q8R5D9 | Gene names | Pdcd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18). | |||||
|
ASPX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.004847 (rank : 159) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.006308 (rank : 149) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.006704 (rank : 146) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.005050 (rank : 158) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
RPA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.003989 (rank : 164) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
SNUT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.006363 (rank : 148) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
SSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.006467 (rank : 147) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQ55, Q9BW97, Q9H170 | Gene names | PPAN, SSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of SWI4 1 homolog (Ssf-1) (Peter Pan homolog). | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.019205 (rank : 117) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.028663 (rank : 108) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.006706 (rank : 145) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.007497 (rank : 142) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
FIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.014828 (rank : 125) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | -0.002808 (rank : 173) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | -0.002296 (rank : 172) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.007911 (rank : 139) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
RUNX3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.002192 (rank : 169) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.018849 (rank : 118) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.057072 (rank : 96) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.015380 (rank : 124) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TR150_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.033272 (rank : 104) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
XPC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.008420 (rank : 138) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | -0.006762 (rank : 176) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CC026_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.161348 (rank : 87) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
CCD55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.053786 (rank : 97) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
FANCM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.386257 (rank : 78) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.382449 (rank : 79) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
HELC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.106758 (rank : 91) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.071203 (rank : 93) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 170 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.995115 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.967656 (rank : 3) | θ value | 4.85792e-54 (rank : 10) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.967349 (rank : 4) | θ value | 1.45929e-58 (rank : 3) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.967147 (rank : 5) | θ value | 4.85792e-54 (rank : 9) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.964015 (rank : 6) | θ value | 2.10721e-57 (rank : 4) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.961595 (rank : 7) | θ value | 5.19021e-56 (rank : 7) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.961150 (rank : 8) | θ value | 4.70527e-49 (rank : 15) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.960976 (rank : 9) | θ value | 4.70527e-49 (rank : 16) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.960058 (rank : 10) | θ value | 4.39379e-55 (rank : 8) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.956946 (rank : 11) | θ value | 3.15612e-45 (rank : 29) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.956833 (rank : 12) | θ value | 1.28137e-46 (rank : 25) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.956460 (rank : 13) | θ value | 3.15612e-45 (rank : 30) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.954895 (rank : 14) | θ value | 1.51363e-47 (rank : 18) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.954827 (rank : 15) | θ value | 6.81017e-40 (rank : 41) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.954803 (rank : 16) | θ value | 5.21438e-40 (rank : 39) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.954551 (rank : 17) | θ value | 5.55816e-50 (rank : 12) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.954027 (rank : 18) | θ value | 1.788e-48 (rank : 17) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.953513 (rank : 19) | θ value | 2.04573e-44 (rank : 33) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.953320 (rank : 20) | θ value | 1.61718e-49 (rank : 13) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.953153 (rank : 21) | θ value | 2.1121e-49 (rank : 14) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.952870 (rank : 22) | θ value | 8.30557e-46 (rank : 28) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.952735 (rank : 23) | θ value | 8.30557e-46 (rank : 27) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.952174 (rank : 24) | θ value | 1.12253e-42 (rank : 38) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.952011 (rank : 25) | θ value | 5.76516e-39 (rank : 42) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.951200 (rank : 26) | θ value | 7.52953e-39 (rank : 45) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.951166 (rank : 27) | θ value | 3.86705e-35 (rank : 52) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.949923 (rank : 28) | θ value | 6.35935e-46 (rank : 26) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.949885 (rank : 29) | θ value | 3.15612e-45 (rank : 31) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.949672 (rank : 30) | θ value | 4.55743e-44 (rank : 34) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.949672 (rank : 31) | θ value | 4.55743e-44 (rank : 35) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.949258 (rank : 32) | θ value | 1.6247e-33 (rank : 54) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.949125 (rank : 33) | θ value | 2.26182e-43 (rank : 36) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.949125 (rank : 34) | θ value | 2.26182e-43 (rank : 37) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.949011 (rank : 35) | θ value | 2.50655e-34 (rank : 53) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.948321 (rank : 36) | θ value | 6.81017e-40 (rank : 40) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.947506 (rank : 37) | θ value | 1.85459e-37 (rank : 47) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.947405 (rank : 38) | θ value | 1.97686e-47 (rank : 19) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.947348 (rank : 39) | θ value | 3.73686e-38 (rank : 46) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.947342 (rank : 40) | θ value | 2.58187e-47 (rank : 20) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.947082 (rank : 41) | θ value | 5.76516e-39 (rank : 43) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.947031 (rank : 42) | θ value | 1.4233e-29 (rank : 65) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.947002 (rank : 43) | θ value | 5.76516e-39 (rank : 44) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.945112 (rank : 44) | θ value | 1.28137e-46 (rank : 24) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.944925 (rank : 45) | θ value | 7.51208e-47 (rank : 23) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.944661 (rank : 46) | θ value | 2.58187e-47 (rank : 21) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.944322 (rank : 47) | θ value | 4.40402e-47 (rank : 22) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.942816 (rank : 48) | θ value | 4.122e-45 (rank : 32) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.942543 (rank : 49) | θ value | 2.32979e-56 (rank : 5) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.940439 (rank : 50) | θ value | 2.32979e-56 (rank : 6) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.938612 (rank : 51) | θ value | 7.04741e-37 (rank : 49) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.938514 (rank : 52) | θ value | 1.57e-36 (rank : 50) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.935633 (rank : 53) | θ value | 1.79631e-32 (rank : 56) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.932224 (rank : 54) | θ value | 7.01481e-53 (rank : 11) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.932194 (rank : 55) | θ value | 2.34606e-32 (rank : 57) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.930346 (rank : 56) | θ value | 3.74554e-30 (rank : 62) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.928695 (rank : 57) | θ value | 1.08979e-29 (rank : 64) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.928588 (rank : 58) | θ value | 4.72714e-33 (rank : 55) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.927540 (rank : 59) | θ value | 8.3442e-30 (rank : 63) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.927428 (rank : 60) | θ value | 3.06405e-32 (rank : 58) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.926269 (rank : 61) | θ value | 7.04741e-37 (rank : 48) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.926148 (rank : 62) | θ value | 8.91499e-32 (rank : 59) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.925540 (rank : 63) | θ value | 1.16434e-31 (rank : 60) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.925442 (rank : 64) | θ value | 1.16434e-31 (rank : 61) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.918506 (rank : 65) | θ value | 1.01765e-35 (rank : 51) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.911196 (rank : 66) | θ value | 3.28125e-26 (rank : 66) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.884089 (rank : 67) | θ value | 3.17815e-21 (rank : 67) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.882427 (rank : 68) | θ value | 7.0802e-21 (rank : 68) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.530729 (rank : 69) | θ value | 0.0148317 (rank : 83) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.517358 (rank : 70) | θ value | 0.00869519 (rank : 77) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.492944 (rank : 71) | θ value | 9.29e-05 (rank : 70) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.479116 (rank : 72) | θ value | 0.000461057 (rank : 72) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.470921 (rank : 73) | θ value | 6.43352e-06 (rank : 69) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.450674 (rank : 74) | θ value | 0.0736092 (rank : 88) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.435904 (rank : 75) | θ value | 0.000602161 (rank : 73) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.423293 (rank : 76) | θ value | 0.0563607 (rank : 86) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.420316 (rank : 77) | θ value | 0.279714 (rank : 93) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.386257 (rank : 78) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.382449 (rank : 79) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.373506 (rank : 80) | θ value | 1.81305 (rank : 109) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.372853 (rank : 81) | θ value | 2.36792 (rank : 115) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.313198 (rank : 82) | θ value | 0.163984 (rank : 90) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.312208 (rank : 83) | θ value | 4.03905 (rank : 138) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.299118 (rank : 84) | θ value | 3.0926 (rank : 125) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.297822 (rank : 85) | θ value | 0.47712 (rank : 95) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.191322 (rank : 86) | θ value | 1.06291 (rank : 103) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.161348 (rank : 87) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.159343 (rank : 88) | θ value | 1.81305 (rank : 108) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.143968 (rank : 89) | θ value | 1.81305 (rank : 114) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
CC026_MOUSE
|
||||||
| NC score | 0.133451 (rank : 90) | θ value | 1.38821 (rank : 104) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.106758 (rank : 91) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
SKIV2_HUMAN
|
||||||
| NC score | 0.087577 (rank : 92) | θ value | 0.0113563 (rank : 81) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.071203 (rank : 93) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
SK2L2_HUMAN
|
||||||
| NC score | 0.066263 (rank : 94) | θ value | 0.0113563 (rank : 79) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42285, Q8TAG2 | Gene names | SKIV2L2, KIAA0052 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
SK2L2_MOUSE
|
||||||
| NC score | 0.066072 (rank : 95) | θ value | 0.0113563 (rank : 80) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CZU3 | Gene names | Skiv2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.057072 (rank : 96) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.053786 (rank : 97) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.052233 (rank : 98) | θ value | 0.00390308 (rank : 76) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.041084 (rank : 99) | θ value | 0.0113563 (rank : 78) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.040703 (rank : 100) | θ value | 0.00020696 (rank : 71) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.040566 (rank : 101) | θ value | 0.0193708 (rank : 84) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
T2_MOUSE
|
||||||
| NC score | 0.040298 (rank : 102) | θ value | 0.163984 (rank : 91) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.037689 (rank : 103) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.033272 (rank : 104) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
SNW1_MOUSE
|
||||||
| NC score | 0.032284 (rank : 105) | θ value | 1.81305 (rank : 113) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CSN1 | Gene names | Snw1, Skiip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNW domain-containing protein 1 (Nuclear protein SkiP) (Ski- interacting protein). | |||||
|
SNW1_HUMAN
|
||||||
| NC score | 0.032214 (rank : 106) | θ value | 1.81305 (rank : 112) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13573, Q13483, Q32N03, Q5D0D6 | Gene names | SNW1, SKIIP, SKIP | |||
|
Domain Architecture |

|
|||||
| Description | SNW domain-containing protein 1 (Nuclear protein SkiP) (Ski- interacting protein) (Nuclear receptor coactivator NCoA-62). | |||||
|
TR150_HUMAN
|
||||||
| NC score | 0.030237 (rank : 107) | θ value | 2.36792 (rank : 121) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.028663 (rank : 108) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.027420 (rank : 109) | θ value | 4.03905 (rank : 142) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
PRP16_HUMAN
|
||||||
| NC score | 0.027397 (rank : 110) | θ value | 0.0148317 (rank : 82) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.027078 (rank : 111) | θ value | 4.03905 (rank : 140) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.025434 (rank : 112) | θ value | 0.0193708 (rank : 85) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.025303 (rank : 113) | θ value | 0.365318 (rank : 94) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.024765 (rank : 114) | θ value | 0.00175202 (rank : 74) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.024623 (rank : 115) | θ value | 0.0563607 (rank : 87) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.024023 (rank : 116) | θ value | 0.00298849 (rank : 75) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.019205 (rank : 117) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.018849 (rank : 118) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
FIP1_HUMAN
|
||||||
| NC score | 0.018454 (rank : 119) | θ value | 3.0926 (rank : 123) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
RU17_HUMAN
|
||||||
| NC score | 0.017443 (rank : 120) | θ value | 0.62314 (rank : 99) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
CT004_MOUSE
|
||||||
| NC score | 0.017290 (rank : 121) | θ value | 0.62314 (rank : 96) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D2V5, Q3TM16, Q8VBU9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf4 homolog. | |||||
|
TCAL6_HUMAN
|
||||||
| NC score | 0.016340 (rank : 122) | θ value | 4.03905 (rank : 141) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6IPX3, Q5H9J8 | Gene names | TCEAL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 6 (TCEA-like protein 6) (Transcription elongation factor S-II protein-like 6). | |||||
|
RU17_MOUSE
|
||||||
| NC score | 0.015965 (rank : 123) | θ value | 2.36792 (rank : 119) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.015380 (rank : 124) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
FIP1_MOUSE
|
||||||
| NC score | 0.014828 (rank : 125) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
EI2BD_MOUSE
|
||||||
| NC score | 0.014826 (rank : 126) | θ value | 0.62314 (rank : 97) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61749, Q61748, Q8VC35 | Gene names | Eif2b4, Eif2b, Eif2bd, Jgr1a | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.014470 (rank : 127) | θ value | 1.38821 (rank : 105) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
RN169_HUMAN
|
||||||
| NC score | 0.013523 (rank : 128) | θ value | 1.81305 (rank : 111) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
HAP1_MOUSE
|
||||||
| NC score | 0.013412 (rank : 129) | θ value | 0.21417 (rank : 92) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35668, O35636 | Gene names | Hap1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1). | |||||
|
WDR60_MOUSE
|
||||||
| NC score | 0.012718 (rank : 130) | θ value | 1.38821 (rank : 107) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.011509 (rank : 131) | θ value | 4.03905 (rank : 134) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
NTHL1_HUMAN
|
||||||
| NC score | 0.009399 (rank : 132) | θ value | 4.03905 (rank : 139) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78549, Q99566, Q99794, Q9BPX2 | Gene names | NTHL1, NTH1, OCTS3 | |||
|
Domain Architecture |

|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
LC7L2_HUMAN
|
||||||
| NC score | 0.008962 (rank : 133) | θ value | 2.36792 (rank : 116) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
LC7L2_MOUSE
|
||||||
| NC score | 0.008915 (rank : 134) | θ value | 3.0926 (rank : 124) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.008518 (rank : 135) | θ value | 0.0961366 (rank : 89) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.008504 (rank : 136) | θ value | 0.813845 (rank : 102) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
COX10_MOUSE
|
||||||
| NC score | 0.008499 (rank : 137) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
XPC_MOUSE
|
||||||
| NC score | 0.008420 (rank : 138) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.007911 (rank : 139) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.007696 (rank : 140) | θ value | 2.36792 (rank : 117) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.007522 (rank : 141) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.007497 (rank : 142) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.006877 (rank : 143) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.006707 (rank : 144) | θ value | 0.62314 (rank : 98) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.006706 (rank : 145) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.006704 (rank : 146) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
SSF1_HUMAN
|
||||||
| NC score | 0.006467 (rank : 147) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQ55, Q9BW97, Q9H170 | Gene names | PPAN, SSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of SWI4 1 homolog (Ssf-1) (Peter Pan homolog). | |||||
|
SNUT1_HUMAN
|
||||||
| NC score | 0.006363 (rank : 148) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.006308 (rank : 149) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
H2BWT_HUMAN
|
||||||
| NC score | 0.006283 (rank : 150) | θ value | 1.38821 (rank : 106) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2G1 | Gene names | H2BFWT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2B type W-T (H2B histone family member W testis-specific). | |||||
|
SAPS2_HUMAN
|
||||||
| NC score | 0.006147 (rank : 151) | θ value | 2.36792 (rank : 120) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
JIP3_MOUSE
|
||||||
| NC score | 0.005942 (rank : 152) | θ value | 4.03905 (rank : 137) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.005774 (rank : 153) | θ value | 3.0926 (rank : 126) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.005624 (rank : 154) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
PDCD7_MOUSE
|
||||||
| NC score | 0.005488 (rank : 155) | θ value | 5.27518 (rank : 148) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTY1, Q8R5D9 | Gene names | Pdcd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18). | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.005310 (rank : 156) | θ value | 4.03905 (rank : 135) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
LBR_MOUSE
|
||||||
| NC score | 0.005104 (rank : 157) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U9G9, Q3TSW2, Q811V8, Q811V9, Q8BST3, Q8K2Y8, Q8VDM0, Q91YS5, Q91Z27 | Gene names | Lbr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.005050 (rank : 158) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
ASPX_HUMAN
|
||||||
| NC score | 0.004847 (rank : 159) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.004275 (rank : 160) | θ value | 0.62314 (rank : 100) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.004077 (rank : 161) | θ value | 4.03905 (rank : 136) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
RING1_MOUSE
|
||||||
| NC score | 0.004053 (rank : 162) | θ value | 2.36792 (rank : 118) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
2A5D_HUMAN
|
||||||
| NC score | 0.004030 (rank : 163) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14738, O00494, O00696, Q15171 | Gene names | PPP2R5D | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (PP2A, B subunit, B' delta isoform) (PP2A, B subunit, B56 delta isoform) (PP2A, B subunit, PR61 delta isoform) (PP2A, B subunit, R5 delta isoform). | |||||
|
RPA1_HUMAN
|
||||||
| NC score | 0.003989 (rank : 164) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.003796 (rank : 165) | θ value | 3.0926 (rank : 130) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.003729 (rank : 166) | θ value | 0.813845 (rank : 101) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.003165 (rank : 167) | θ value | 3.0926 (rank : 129) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.002911 (rank : 168) | θ value | 3.0926 (rank : 122) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RUNX3_MOUSE
|
||||||
| NC score | 0.002192 (rank : 169) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.001749 (rank : 170) | θ value | 3.0926 (rank : 127) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.000723 (rank : 171) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | -0.002296 (rank : 172) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MACF1_HUMAN
|
||||||
| NC score | -0.002808 (rank : 173) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | -0.004109 (rank : 174) | θ value | 3.0926 (rank : 128) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | -0.004901 (rank : 175) | θ value | 1.81305 (rank : 110) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | -0.006762 (rank : 176) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||