Please be patient as the page loads
|
DDX55_HUMAN
|
||||||
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DDX55_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998157 (rank : 2) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 2.31364e-80 (rank : 3) | NC score | 0.970416 (rank : 5) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 5.15425e-80 (rank : 4) | NC score | 0.969463 (rank : 6) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | 2.09745e-73 (rank : 5) | NC score | 0.975794 (rank : 4) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 6.10265e-73 (rank : 6) | NC score | 0.977421 (rank : 3) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 5.19021e-56 (rank : 7) | NC score | 0.966188 (rank : 9) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 1.32293e-51 (rank : 8) | NC score | 0.967139 (rank : 8) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 5.02714e-51 (rank : 9) | NC score | 0.967504 (rank : 7) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 2.41655e-45 (rank : 10) | NC score | 0.955166 (rank : 15) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 2.41655e-45 (rank : 11) | NC score | 0.954985 (rank : 17) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 4.122e-45 (rank : 12) | NC score | 0.956537 (rank : 12) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 5.38352e-45 (rank : 13) | NC score | 0.956283 (rank : 13) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 1.56636e-44 (rank : 14) | NC score | 0.925917 (rank : 59) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 1.01529e-43 (rank : 15) | NC score | 0.932918 (rank : 53) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 1.01529e-43 (rank : 16) | NC score | 0.930511 (rank : 55) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 17) | NC score | 0.947411 (rank : 29) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 3.85808e-43 (rank : 18) | NC score | 0.947547 (rank : 28) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 3.61106e-41 (rank : 19) | NC score | 0.952737 (rank : 19) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 2.34062e-40 (rank : 20) | NC score | 0.951411 (rank : 21) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 3.99248e-40 (rank : 21) | NC score | 0.950407 (rank : 24) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 1.16164e-39 (rank : 22) | NC score | 0.954559 (rank : 18) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 23) | NC score | 0.955415 (rank : 14) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 24) | NC score | 0.957028 (rank : 11) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 5.76516e-39 (rank : 25) | NC score | 0.943695 (rank : 33) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 26) | NC score | 0.943524 (rank : 35) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 27) | NC score | 0.951200 (rank : 22) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 28) | NC score | 0.942882 (rank : 38) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 9.83387e-39 (rank : 29) | NC score | 0.942743 (rank : 39) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 2.86122e-38 (rank : 30) | NC score | 0.936640 (rank : 46) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 31) | NC score | 0.936287 (rank : 47) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 1.42001e-37 (rank : 32) | NC score | 0.935587 (rank : 50) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 1.20211e-36 (rank : 33) | NC score | 0.934452 (rank : 52) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 1.20211e-36 (rank : 34) | NC score | 0.955094 (rank : 16) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 1.57e-36 (rank : 35) | NC score | 0.958890 (rank : 10) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 1.57e-36 (rank : 36) | NC score | 0.942030 (rank : 40) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 1.57e-36 (rank : 37) | NC score | 0.942030 (rank : 41) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 1.57e-36 (rank : 38) | NC score | 0.941631 (rank : 42) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 1.57e-36 (rank : 39) | NC score | 0.941631 (rank : 43) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 2.05049e-36 (rank : 40) | NC score | 0.937123 (rank : 45) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
UAP56_HUMAN
|
||||||
| θ value | 4.568e-36 (rank : 41) | NC score | 0.943371 (rank : 36) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | 4.568e-36 (rank : 42) | NC score | 0.943290 (rank : 37) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | 7.79185e-36 (rank : 43) | NC score | 0.943551 (rank : 34) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 1.01765e-35 (rank : 44) | NC score | 0.946487 (rank : 30) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 45) | NC score | 0.944065 (rank : 32) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 8.61488e-35 (rank : 46) | NC score | 0.926014 (rank : 58) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 7.29293e-34 (rank : 47) | NC score | 0.918309 (rank : 61) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 48) | NC score | 0.949861 (rank : 26) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 8.06329e-33 (rank : 49) | NC score | 0.950238 (rank : 25) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | 3.06405e-32 (rank : 50) | NC score | 0.950886 (rank : 23) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 4.00176e-32 (rank : 51) | NC score | 0.935962 (rank : 48) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 52) | NC score | 0.939853 (rank : 44) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX56_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 53) | NC score | 0.951447 (rank : 20) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 3.3877e-31 (rank : 54) | NC score | 0.935614 (rank : 49) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 9.8567e-31 (rank : 55) | NC score | 0.934467 (rank : 51) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 56) | NC score | 0.922563 (rank : 60) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 57) | NC score | 0.931458 (rank : 54) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 2.68423e-28 (rank : 58) | NC score | 0.947627 (rank : 27) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 59) | NC score | 0.917639 (rank : 63) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 60) | NC score | 0.929433 (rank : 56) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 61) | NC score | 0.926134 (rank : 57) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 62) | NC score | 0.918211 (rank : 62) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 63) | NC score | 0.944884 (rank : 31) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 64) | NC score | 0.916935 (rank : 66) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 65) | NC score | 0.917120 (rank : 64) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 66) | NC score | 0.917010 (rank : 65) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 67) | NC score | 0.871096 (rank : 68) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 68) | NC score | 0.871858 (rank : 67) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 69) | NC score | 0.420278 (rank : 75) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
U520_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 70) | NC score | 0.159881 (rank : 88) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 71) | NC score | 0.409618 (rank : 78) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
DICER_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 72) | NC score | 0.410851 (rank : 77) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 73) | NC score | 0.325705 (rank : 82) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
NPHP3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 74) | NC score | 0.030445 (rank : 98) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 75) | NC score | 0.043614 (rank : 93) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
NPHP3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 76) | NC score | 0.030739 (rank : 97) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 77) | NC score | 0.503655 (rank : 69) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 78) | NC score | 0.309316 (rank : 84) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 79) | NC score | 0.420822 (rank : 74) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 80) | NC score | 0.037914 (rank : 94) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 0.125558 (rank : 81) | NC score | 0.421830 (rank : 73) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 82) | NC score | 0.485272 (rank : 70) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
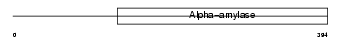
4F2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 83) | NC score | 0.029946 (rank : 99) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08195, Q13543 | Gene names | SLC3A2, MDU1 | |||
|
Domain Architecture |
|
|||||
| Description | 4F2 cell-surface antigen heavy chain (4F2hc) (Lymphocyte activation antigen 4F2 large subunit) (4F2 heavy chain antigen) (CD98 antigen). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 84) | NC score | 0.010704 (rank : 104) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
PZRN3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 85) | NC score | 0.008355 (rank : 107) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 86) | NC score | 0.015829 (rank : 100) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 87) | NC score | 0.322170 (rank : 83) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.008236 (rank : 108) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.015576 (rank : 101) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.307850 (rank : 85) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.005636 (rank : 115) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.003450 (rank : 119) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.377628 (rank : 79) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
K0020_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.008623 (rank : 106) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKS9, Q6A0E6, Q8BU15 | Gene names | Kiaa0020 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0020. | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | -0.001515 (rank : 128) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.003654 (rank : 117) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
P85B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.003473 (rank : 118) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
IMPG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.012188 (rank : 102) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1W8, Q9CTP8, Q9ES62, Q9ET31 | Gene names | Impg1, Spacr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interphotoreceptor matrix proteoglycan 1 precursor (IPM 150 proteoglycan) (Sialoprotein associated with cones and rods). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.000018 (rank : 126) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
WRN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.411766 (rank : 76) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
BLM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.427853 (rank : 72) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.203634 (rank : 86) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.003315 (rank : 121) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
NFIA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.006855 (rank : 113) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12857, Q9H3X9, Q9P2A9 | Gene names | NFIA, KIAA1439 | |||
|
Domain Architecture |
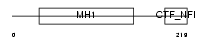
|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NFIA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.006966 (rank : 112) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02780, P70250, P70251, Q3UUZ2, Q61960, Q8VBT5, Q9R1G5 | Gene names | Nfia | |||
|
Domain Architecture |
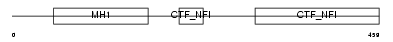
|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NTBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.007354 (rank : 110) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.006772 (rank : 114) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
DDX58_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.172212 (rank : 87) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.004601 (rank : 116) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
P85B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.003114 (rank : 122) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | -0.004118 (rank : 129) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.001019 (rank : 125) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.035590 (rank : 95) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.035381 (rank : 96) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CP46A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | -0.001164 (rank : 127) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6A2 | Gene names | CYP46A1, CYP46 | |||
|
Domain Architecture |
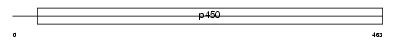
|
|||||
| Description | Cytochrome p450 46A1 (EC 1.14.13.98) (Cholesterol 24-hydroxylase) (CH24H). | |||||
|
ERCC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.003364 (rank : 120) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
HSP7C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.001202 (rank : 123) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.001191 (rank : 124) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
PDZK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.010726 (rank : 103) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q76G19, Q8NB75, Q9BUH9, Q9P284 | Gene names | PDZK4, KIAA1444, PDZRN4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
PDZK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.010668 (rank : 105) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QY39, Q6PHP2 | Gene names | Pdzk4, Pdzrn4l, Xlu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
SPF30_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.007343 (rank : 111) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75940 | Gene names | SMNDC1, SMNR, SPF30 | |||
|
Domain Architecture |
|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
SPF30_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.007387 (rank : 109) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGT7, Q4QQL0 | Gene names | Smndc1, Smnr, Spf30 | |||
|
Domain Architecture |
|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
BLM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.438439 (rank : 71) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
CC026_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.153137 (rank : 89) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
CC026_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.121556 (rank : 90) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
HELC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.106870 (rank : 91) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
RECQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.375998 (rank : 80) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.071557 (rank : 92) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
WRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.373745 (rank : 81) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.998157 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.977421 (rank : 3) | θ value | 6.10265e-73 (rank : 6) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.975794 (rank : 4) | θ value | 2.09745e-73 (rank : 5) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.970416 (rank : 5) | θ value | 2.31364e-80 (rank : 3) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.969463 (rank : 6) | θ value | 5.15425e-80 (rank : 4) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.967504 (rank : 7) | θ value | 5.02714e-51 (rank : 9) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.967139 (rank : 8) | θ value | 1.32293e-51 (rank : 8) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.966188 (rank : 9) | θ value | 5.19021e-56 (rank : 7) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.958890 (rank : 10) | θ value | 1.57e-36 (rank : 35) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.957028 (rank : 11) | θ value | 3.37985e-39 (rank : 24) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.956537 (rank : 12) | θ value | 4.122e-45 (rank : 12) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.956283 (rank : 13) | θ value | 5.38352e-45 (rank : 13) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.955415 (rank : 14) | θ value | 1.98146e-39 (rank : 23) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.955166 (rank : 15) | θ value | 2.41655e-45 (rank : 10) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.955094 (rank : 16) | θ value | 1.20211e-36 (rank : 34) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.954985 (rank : 17) | θ value | 2.41655e-45 (rank : 11) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.954559 (rank : 18) | θ value | 1.16164e-39 (rank : 22) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.952737 (rank : 19) | θ value | 3.61106e-41 (rank : 19) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.951447 (rank : 20) | θ value | 1.52067e-31 (rank : 53) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.951411 (rank : 21) | θ value | 2.34062e-40 (rank : 20) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.951200 (rank : 22) | θ value | 7.52953e-39 (rank : 27) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.950886 (rank : 23) | θ value | 3.06405e-32 (rank : 50) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.950407 (rank : 24) | θ value | 3.99248e-40 (rank : 21) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.950238 (rank : 25) | θ value | 8.06329e-33 (rank : 49) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.949861 (rank : 26) | θ value | 8.06329e-33 (rank : 48) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.947627 (rank : 27) | θ value | 2.68423e-28 (rank : 58) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.947547 (rank : 28) | θ value | 3.85808e-43 (rank : 18) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.947411 (rank : 29) | θ value | 3.85808e-43 (rank : 17) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.946487 (rank : 30) | θ value | 1.01765e-35 (rank : 44) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.944884 (rank : 31) | θ value | 1.12775e-26 (rank : 63) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.944065 (rank : 32) | θ value | 1.32909e-35 (rank : 45) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.943695 (rank : 33) | θ value | 5.76516e-39 (rank : 25) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.943551 (rank : 34) | θ value | 7.79185e-36 (rank : 43) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.943524 (rank : 35) | θ value | 7.52953e-39 (rank : 26) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.943371 (rank : 36) | θ value | 4.568e-36 (rank : 41) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.943290 (rank : 37) | θ value | 4.568e-36 (rank : 42) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.942882 (rank : 38) | θ value | 7.52953e-39 (rank : 28) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.942743 (rank : 39) | θ value | 9.83387e-39 (rank : 29) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.942030 (rank : 40) | θ value | 1.57e-36 (rank : 36) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.942030 (rank : 41) | θ value | 1.57e-36 (rank : 37) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.941631 (rank : 42) | θ value | 1.57e-36 (rank : 38) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.941631 (rank : 43) | θ value | 1.57e-36 (rank : 39) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.939853 (rank : 44) | θ value | 5.22648e-32 (rank : 52) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.937123 (rank : 45) | θ value | 2.05049e-36 (rank : 40) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.936640 (rank : 46) | θ value | 2.86122e-38 (rank : 30) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.936287 (rank : 47) | θ value | 4.88048e-38 (rank : 31) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.935962 (rank : 48) | θ value | 4.00176e-32 (rank : 51) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.935614 (rank : 49) | θ value | 3.3877e-31 (rank : 54) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.935587 (rank : 50) | θ value | 1.42001e-37 (rank : 32) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.934467 (rank : 51) | θ value | 9.8567e-31 (rank : 55) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.934452 (rank : 52) | θ value | 1.20211e-36 (rank : 33) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.932918 (rank : 53) | θ value | 1.01529e-43 (rank : 15) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.931458 (rank : 54) | θ value | 1.08979e-29 (rank : 57) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.930511 (rank : 55) | θ value | 1.01529e-43 (rank : 16) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.929433 (rank : 56) | θ value | 1.73987e-27 (rank : 60) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.926134 (rank : 57) | θ value | 1.73987e-27 (rank : 61) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.926014 (rank : 58) | θ value | 8.61488e-35 (rank : 46) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.925917 (rank : 59) | θ value | 1.56636e-44 (rank : 14) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.922563 (rank : 60) | θ value | 9.8567e-31 (rank : 56) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.918309 (rank : 61) | θ value | 7.29293e-34 (rank : 47) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.918211 (rank : 62) | θ value | 2.27234e-27 (rank : 62) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.917639 (rank : 63) | θ value | 1.73987e-27 (rank : 59) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.917120 (rank : 64) | θ value | 7.56453e-23 (rank : 65) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.917010 (rank : 65) | θ value | 4.15078e-21 (rank : 66) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.916935 (rank : 66) | θ value | 7.30988e-26 (rank : 64) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.871858 (rank : 67) | θ value | 6.20254e-17 (rank : 68) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.871096 (rank : 68) | θ value | 2.7842e-17 (rank : 67) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.503655 (rank : 69) | θ value | 0.0252991 (rank : 77) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.485272 (rank : 70) | θ value | 0.21417 (rank : 82) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.438439 (rank : 71) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.427853 (rank : 72) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.421830 (rank : 73) | θ value | 0.125558 (rank : 81) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.420822 (rank : 74) | θ value | 0.0431538 (rank : 79) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.420278 (rank : 75) | θ value | 0.000461057 (rank : 69) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.411766 (rank : 76) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.410851 (rank : 77) | θ value | 0.00102713 (rank : 72) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.409618 (rank : 78) | θ value | 0.000786445 (rank : 71) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.377628 (rank : 79) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.375998 (rank : 80) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.373745 (rank : 81) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.325705 (rank : 82) | θ value | 0.00509761 (rank : 73) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.322170 (rank : 83) | θ value | 1.06291 (rank : 87) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.309316 (rank : 84) | θ value | 0.0330416 (rank : 78) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.307850 (rank : 85) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.203634 (rank : 86) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.172212 (rank : 87) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.159881 (rank : 88) | θ value | 0.000602161 (rank : 70) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.153137 (rank : 89) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
CC026_MOUSE
|
||||||
| NC score | 0.121556 (rank : 90) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.106870 (rank : 91) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.071557 (rank : 92) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.043614 (rank : 93) | θ value | 0.0148317 (rank : 75) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.037914 (rank : 94) | θ value | 0.0563607 (rank : 80) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.035590 (rank : 95) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.035381 (rank : 96) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
NPHP3_MOUSE
|
||||||
| NC score | 0.030739 (rank : 97) | θ value | 0.0252991 (rank : 76) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
NPHP3_HUMAN
|
||||||
| NC score | 0.030445 (rank : 98) | θ value | 0.00869519 (rank : 74) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
4F2_HUMAN
|
||||||
| NC score | 0.029946 (rank : 99) | θ value | 0.62314 (rank : 83) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08195, Q13543 | Gene names | SLC3A2, MDU1 | |||
|
Domain Architecture |

|
|||||
| Description | 4F2 cell-surface antigen heavy chain (4F2hc) (Lymphocyte activation antigen 4F2 large subunit) (4F2 heavy chain antigen) (CD98 antigen). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.015829 (rank : 100) | θ value | 1.06291 (rank : 86) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.015576 (rank : 101) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
IMPG1_MOUSE
|
||||||
| NC score | 0.012188 (rank : 102) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1W8, Q9CTP8, Q9ES62, Q9ET31 | Gene names | Impg1, Spacr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interphotoreceptor matrix proteoglycan 1 precursor (IPM 150 proteoglycan) (Sialoprotein associated with cones and rods). | |||||
|
PDZK4_HUMAN
|
||||||
| NC score | 0.010726 (rank : 103) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q76G19, Q8NB75, Q9BUH9, Q9P284 | Gene names | PDZK4, KIAA1444, PDZRN4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.010704 (rank : 104) | θ value | 0.62314 (rank : 84) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
PDZK4_MOUSE
|
||||||
| NC score | 0.010668 (rank : 105) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QY39, Q6PHP2 | Gene names | Pdzk4, Pdzrn4l, Xlu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
K0020_MOUSE
|
||||||
| NC score | 0.008623 (rank : 106) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKS9, Q6A0E6, Q8BU15 | Gene names | Kiaa0020 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0020. | |||||
|
PZRN3_MOUSE
|
||||||
| NC score | 0.008355 (rank : 107) | θ value | 0.813845 (rank : 85) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.008236 (rank : 108) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
SPF30_MOUSE
|
||||||
| NC score | 0.007387 (rank : 109) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGT7, Q4QQL0 | Gene names | Smndc1, Smnr, Spf30 | |||
|
Domain Architecture |

|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
NTBP1_MOUSE
|
||||||
| NC score | 0.007354 (rank : 110) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
SPF30_HUMAN
|
||||||
| NC score | 0.007343 (rank : 111) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75940 | Gene names | SMNDC1, SMNR, SPF30 | |||
|
Domain Architecture |

|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
NFIA_MOUSE
|
||||||
| NC score | 0.006966 (rank : 112) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02780, P70250, P70251, Q3UUZ2, Q61960, Q8VBT5, Q9R1G5 | Gene names | Nfia | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NFIA_HUMAN
|
||||||
| NC score | 0.006855 (rank : 113) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12857, Q9H3X9, Q9P2A9 | Gene names | NFIA, KIAA1439 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.006772 (rank : 114) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.005636 (rank : 115) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.004601 (rank : 116) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.003654 (rank : 117) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.003473 (rank : 118) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.003450 (rank : 119) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
ERCC2_HUMAN
|
||||||
| NC score | 0.003364 (rank : 120) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.003315 (rank : 121) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.003114 (rank : 122) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
HSP7C_HUMAN
|
||||||
| NC score | 0.001202 (rank : 123) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| NC score | 0.001191 (rank : 124) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.001019 (rank : 125) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.000018 (rank : 126) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
CP46A_HUMAN
|
||||||
| NC score | -0.001164 (rank : 127) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6A2 | Gene names | CYP46A1, CYP46 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome p450 46A1 (EC 1.14.13.98) (Cholesterol 24-hydroxylase) (CH24H). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | -0.001515 (rank : 128) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
STK10_MOUSE
|
||||||
| NC score | -0.004118 (rank : 129) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||