Please be patient as the page loads
|
DICER_MOUSE
|
||||||
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DICER_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994583 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
DDX58_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 3) | NC score | 0.587644 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | 1.38178e-16 (rank : 4) | NC score | 0.565354 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 5) | NC score | 0.588540 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 6) | NC score | 0.573400 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 7) | NC score | 0.580286 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | 2.0648e-12 (rank : 8) | NC score | 0.574109 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 9) | NC score | 0.494386 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
RNC_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 10) | NC score | 0.271927 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 11) | NC score | 0.487765 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 12) | NC score | 0.430701 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 13) | NC score | 0.430552 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 14) | NC score | 0.431607 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 15) | NC score | 0.430860 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 16) | NC score | 0.429178 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 17) | NC score | 0.428984 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 18) | NC score | 0.432829 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 19) | NC score | 0.432829 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 20) | NC score | 0.432620 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 21) | NC score | 0.432620 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 22) | NC score | 0.430795 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 23) | NC score | 0.430798 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 24) | NC score | 0.424975 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 25) | NC score | 0.427071 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 26) | NC score | 0.427412 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 27) | NC score | 0.421117 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 28) | NC score | 0.425101 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 29) | NC score | 0.415306 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 30) | NC score | 0.409618 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 31) | NC score | 0.409222 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 32) | NC score | 0.401518 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 33) | NC score | 0.416583 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 34) | NC score | 0.416609 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
U520_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 35) | NC score | 0.206236 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 36) | NC score | 0.409351 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 37) | NC score | 0.415346 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 38) | NC score | 0.418611 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 39) | NC score | 0.416874 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 40) | NC score | 0.420450 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 41) | NC score | 0.411599 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 42) | NC score | 0.410566 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 43) | NC score | 0.418579 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 44) | NC score | 0.395905 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 45) | NC score | 0.397431 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 46) | NC score | 0.419207 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 47) | NC score | 0.412604 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 48) | NC score | 0.412673 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 49) | NC score | 0.402259 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 50) | NC score | 0.403720 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 51) | NC score | 0.409479 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 52) | NC score | 0.397817 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
HELC1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 53) | NC score | 0.201965 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
PIWL1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 54) | NC score | 0.097419 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96J94, O95404, Q8NA60, Q8TBY5, Q96JD5 | Gene names | PIWIL1, HIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
PIWL1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 55) | NC score | 0.096706 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMB7 | Gene names | Piwil1, Miwi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 56) | NC score | 0.388875 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 57) | NC score | 0.387674 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 58) | NC score | 0.397450 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 59) | NC score | 0.384227 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 60) | NC score | 0.292864 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 61) | NC score | 0.406027 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 62) | NC score | 0.406034 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 63) | NC score | 0.404950 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 64) | NC score | 0.400558 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 65) | NC score | 0.399124 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 66) | NC score | 0.391180 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 67) | NC score | 0.395611 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 68) | NC score | 0.394564 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 69) | NC score | 0.390612 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 70) | NC score | 0.390350 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 71) | NC score | 0.402038 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 72) | NC score | 0.391323 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
TBL2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 73) | NC score | 0.045026 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R099 | Gene names | Tbl2 | |||
|
Domain Architecture |
|
|||||
| Description | Transducin beta-like 2 protein. | |||||
|
UAP56_HUMAN
|
||||||
| θ value | 0.125558 (rank : 74) | NC score | 0.389138 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | 0.125558 (rank : 75) | NC score | 0.389090 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 76) | NC score | 0.373464 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 77) | NC score | 0.374877 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 0.163984 (rank : 78) | NC score | 0.395632 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
PIWL4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 79) | NC score | 0.087156 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGT6, Q8CC75 | Gene names | Piwil4, Miwi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 4 (mAgo5). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 80) | NC score | 0.269246 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | 0.21417 (rank : 81) | NC score | 0.383325 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
ECM29_MOUSE
|
||||||
| θ value | 0.21417 (rank : 82) | NC score | 0.056071 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 0.47712 (rank : 83) | NC score | 0.378483 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
PIWL4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 84) | NC score | 0.077912 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z3Z4, Q68CZ4, Q8N8G9, Q8N9V8, Q8NEH2 | Gene names | PIWIL4, HILI2, PIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 4. | |||||
|
RECQ1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 85) | NC score | 0.208175 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 0.62314 (rank : 86) | NC score | 0.390467 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 0.62314 (rank : 87) | NC score | 0.376487 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 88) | NC score | 0.023961 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
K1914_HUMAN
|
||||||
| θ value | 0.62314 (rank : 89) | NC score | 0.043860 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
AN32E_HUMAN
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.028267 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
AN32A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 91) | NC score | 0.028493 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| θ value | 1.06291 (rank : 92) | NC score | 0.028493 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
TICN2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 93) | NC score | 0.018253 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
ADA19_HUMAN
|
||||||
| θ value | 1.38821 (rank : 94) | NC score | 0.009490 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 95) | NC score | 0.027236 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | 0.021299 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
GNL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 97) | NC score | 0.035027 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36916 | Gene names | Gnl1, Gna-rs1, Mmr1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein MMR1). | |||||
|
PIWL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 98) | NC score | 0.071316 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CDG1, Q3TQE8, Q99MV6, Q9JMB6 | Gene names | Piwil2, Mili | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.012272 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PIWL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.069318 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TC59, Q96SW6, Q9NW28 | Gene names | PIWIL2, HILI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.372853 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.001363 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.014220 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PPM1F_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.018336 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.016216 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
PTPRD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.000888 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
SUHW3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.001915 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
TSP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.009235 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35443, Q86TG2 | Gene names | THBS4, TSP4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
TSP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.009246 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1T2, Q9QYS3, Q9WUE0 | Gene names | Thbs4, Tsp4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
GNL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.029358 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36915 | Gene names | GNL1, HSR1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein HSR1). | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.015132 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
K1914_MOUSE
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.041902 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
M3K6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | -0.003421 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95382 | Gene names | MAP3K6, MAPKKK6 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 6 (EC 2.7.11.25). | |||||
|
MRC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.012501 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.195917 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
ZBTB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | -0.000713 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VL9, Q8CDP7, Q99LD2 | Gene names | Zbtb1 | |||
|
Domain Architecture |
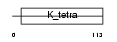
|
|||||
| Description | Zinc finger and BTB domain-containing protein 1. | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.002113 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ENW1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.025836 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQF0, O95244, O95245, Q8NHY7, Q9NRZ2, Q9NZG3 | Gene names | ERVWE1 | |||
|
Domain Architecture |
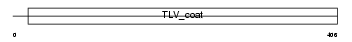
|
|||||
| Description | HERV-W_7q21.2 provirus ancestral Env polyprotein precursor (Envelope polyprotein gPr73) (HERV-7q Envelope protein) (HERV-W envelope protein) (Syncytin) (Syncytin-1) (Enverin) (Env-W) [Contains: Surface protein (SU) (gp50); Transmembrane protein (TM) (gp24)]. | |||||
|
HERC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.014926 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
PRELP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.002554 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JK53 | Gene names | Prelp | |||
|
Domain Architecture |

|
|||||
| Description | Prolargin precursor (Proline-arginine-rich end leucine-rich repeat protein). | |||||
|
UBE2W_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.011801 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDW4, Q9D5H3 | Gene names | Ube2w | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin-conjugating enzyme E2 W (EC 6.3.2.19) (Ubiquitin- protein ligase W) (Ubiquitin carrier protein W). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.000870 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.004803 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
KLDC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.005307 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFQ9, Q6ZPT4 | Gene names | Klhdc5, Kiaa1340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 5. | |||||
|
MRC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.010305 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
PDIA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.006464 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08003, P15841 | Gene names | Pdia4, Cai, Erp72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
UBE2W_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.010727 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96B02, Q9H823, Q9HAG6, Q9NV07 | Gene names | UBE2W | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin-conjugating enzyme E2 W (EC 6.3.2.19) (Ubiquitin- protein ligase W) (Ubiquitin carrier protein W). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.190066 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.011104 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.000395 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
APBB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.006762 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
CLCN5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.004428 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVD4 | Gene names | Clcn5, Clc5 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 5 (ClC-5). | |||||
|
DUS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.005572 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
ISY1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.020996 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULR0, Q96IL2, Q9BT05 | Gene names | KIAA1160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor ISY1 homolog. | |||||
|
ISY1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.021316 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q69ZQ2, Q8VDX4, Q9D3E2 | Gene names | Kiaa1160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor ISY1 homolog. | |||||
|
MRE11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.011543 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49959, O43475 | Gene names | MRE11A, HNGS1, MRE11 | |||
|
Domain Architecture |
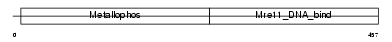
|
|||||
| Description | Double-strand break repair protein MRE11A (MRE11 homolog 1) (MRE11 meiotic recombination 11 homolog A). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.011537 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
PARP9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.008045 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |
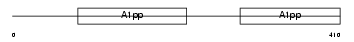
|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
TBL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.017218 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4P3, Q9UQE2 | Gene names | TBL2, WBSCR13 | |||
|
Domain Architecture |
|
|||||
| Description | Transducin beta-like 2 protein (WS beta-transducin repeats protein) (WS-betaTRP) (Williams-Beuren syndrome chromosome region 13 protein). | |||||
|
WRN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.201951 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
BLM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.189853 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
BLM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.194726 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
CC026_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.062842 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
DDX56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.352376 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.352759 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
PIWL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.053937 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z3Z3 | Gene names | PIWIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 3. | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.194362 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.994583 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.588540 (rank : 3) | θ value | 1.80466e-16 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.587644 (rank : 4) | θ value | 1.37858e-24 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.580286 (rank : 5) | θ value | 2.60593e-15 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.574109 (rank : 6) | θ value | 2.0648e-12 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.573400 (rank : 7) | θ value | 1.52774e-15 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.565354 (rank : 8) | θ value | 1.38178e-16 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.494386 (rank : 9) | θ value | 1.9326e-10 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.487765 (rank : 10) | θ value | 6.21693e-09 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.432829 (rank : 11) | θ value | 2.44474e-05 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.432829 (rank : 12) | θ value | 2.44474e-05 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.432620 (rank : 13) | θ value | 4.1701e-05 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.432620 (rank : 14) | θ value | 4.1701e-05 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.431607 (rank : 15) | θ value | 4.92598e-06 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.430860 (rank : 16) | θ value | 8.40245e-06 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.430798 (rank : 17) | θ value | 5.44631e-05 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.430795 (rank : 18) | θ value | 5.44631e-05 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.430701 (rank : 19) | θ value | 2.21117e-06 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.430552 (rank : 20) | θ value | 2.21117e-06 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.429178 (rank : 21) | θ value | 8.40245e-06 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.428984 (rank : 22) | θ value | 1.87187e-05 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.427412 (rank : 23) | θ value | 0.000158464 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.427071 (rank : 24) | θ value | 7.1131e-05 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.425101 (rank : 25) | θ value | 0.000270298 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.424975 (rank : 26) | θ value | 7.1131e-05 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.421117 (rank : 27) | θ value | 0.00020696 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.420450 (rank : 28) | θ value | 0.00134147 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.419207 (rank : 29) | θ value | 0.00390308 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.418611 (rank : 30) | θ value | 0.00134147 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.418579 (rank : 31) | θ value | 0.00228821 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.416874 (rank : 32) | θ value | 0.00134147 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.416609 (rank : 33) | θ value | 0.00102713 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.416583 (rank : 34) | θ value | 0.00102713 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.415346 (rank : 35) | θ value | 0.00134147 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.415306 (rank : 36) | θ value | 0.000786445 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.412673 (rank : 37) | θ value | 0.00509761 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.412604 (rank : 38) | θ value | 0.00509761 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.411599 (rank : 39) | θ value | 0.00175202 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.410566 (rank : 40) | θ value | 0.00175202 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.409618 (rank : 41) | θ value | 0.000786445 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.409479 (rank : 42) | θ value | 0.00509761 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.409351 (rank : 43) | θ value | 0.00134147 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.409222 (rank : 44) | θ value | 0.000786445 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.406034 (rank : 45) | θ value | 0.0252991 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.406027 (rank : 46) | θ value | 0.0252991 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.404950 (rank : 47) | θ value | 0.0330416 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.403720 (rank : 48) | θ value | 0.00509761 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.402259 (rank : 49) | θ value | 0.00509761 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.402038 (rank : 50) | θ value | 0.0736092 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.401518 (rank : 51) | θ value | 0.00102713 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.400558 (rank : 52) | θ value | 0.0330416 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.399124 (rank : 53) | θ value | 0.0330416 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.397817 (rank : 54) | θ value | 0.00665767 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.397450 (rank : 55) | θ value | 0.0148317 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.397431 (rank : 56) | θ value | 0.00390308 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.395905 (rank : 57) | θ value | 0.00298849 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.395632 (rank : 58) | θ value | 0.163984 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.395611 (rank : 59) | θ value | 0.0431538 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.394564 (rank : 60) | θ value | 0.0431538 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.391323 (rank : 61) | θ value | 0.0961366 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.391180 (rank : 62) | θ value | 0.0431538 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.390612 (rank : 63) | θ value | 0.0563607 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.390467 (rank : 64) | θ value | 0.62314 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.390350 (rank : 65) | θ value | 0.0563607 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.389138 (rank : 66) | θ value | 0.125558 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.389090 (rank : 67) | θ value | 0.125558 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.388875 (rank : 68) | θ value | 0.0113563 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.387674 (rank : 69) | θ value | 0.0113563 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.384227 (rank : 70) | θ value | 0.0148317 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.383325 (rank : 71) | θ value | 0.21417 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.378483 (rank : 72) | θ value | 0.47712 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.376487 (rank : 73) | θ value | 0.62314 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.374877 (rank : 74) | θ value | 0.163984 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.373464 (rank : 75) | θ value | 0.163984 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.372853 (rank : 76) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.352759 (rank : 77) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.352376 (rank : 78) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.292864 (rank : 79) | θ value | 0.0193708 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.271927 (rank : 80) | θ value | 1.25267e-09 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.269246 (rank : 81) | θ value | 0.163984 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.208175 (rank : 82) | θ value | 0.47712 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.206236 (rank : 83) | θ value | 0.00102713 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.201965 (rank : 84) | θ value | 0.00665767 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.201951 (rank : 85) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.195917 (rank : 86) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.194726 (rank : 87) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.194362 (rank : 88) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.190066 (rank : 89) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.189853 (rank : 90) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
PIWL1_HUMAN
|
||||||
| NC score | 0.097419 (rank : 91) | θ value | 0.00665767 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96J94, O95404, Q8NA60, Q8TBY5, Q96JD5 | Gene names | PIWIL1, HIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
PIWL1_MOUSE
|
||||||
| NC score | 0.096706 (rank : 92) | θ value | 0.00869519 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMB7 | Gene names | Piwil1, Miwi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
PIWL4_MOUSE
|
||||||
| NC score | 0.087156 (rank : 93) | θ value | 0.163984 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGT6, Q8CC75 | Gene names | Piwil4, Miwi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 4 (mAgo5). | |||||
|
PIWL4_HUMAN
|
||||||
| NC score | 0.077912 (rank : 94) | θ value | 0.47712 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z3Z4, Q68CZ4, Q8N8G9, Q8N9V8, Q8NEH2 | Gene names | PIWIL4, HILI2, PIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 4. | |||||
|
PIWL2_MOUSE
|
||||||
| NC score | 0.071316 (rank : 95) | θ value | 1.38821 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CDG1, Q3TQE8, Q99MV6, Q9JMB6 | Gene names | Piwil2, Mili | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
PIWL2_HUMAN
|
||||||
| NC score | 0.069318 (rank : 96) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TC59, Q96SW6, Q9NW28 | Gene names | PIWIL2, HILI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.062842 (rank : 97) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
ECM29_MOUSE
|
||||||
| NC score | 0.056071 (rank : 98) | θ value | 0.21417 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
PIWL3_HUMAN
|
||||||
| NC score | 0.053937 (rank : 99) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z3Z3 | Gene names | PIWIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 3. | |||||
|
TBL2_MOUSE
|
||||||
| NC score | 0.045026 (rank : 100) | θ value | 0.0961366 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R099 | Gene names | Tbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin beta-like 2 protein. | |||||
|
K1914_HUMAN
|
||||||
| NC score | 0.043860 (rank : 101) | θ value | 0.62314 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
K1914_MOUSE
|
||||||
| NC score | 0.041902 (rank : 102) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
GNL1_MOUSE
|
||||||
| NC score | 0.035027 (rank : 103) | θ value | 1.38821 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36916 | Gene names | Gnl1, Gna-rs1, Mmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein MMR1). | |||||
|
GNL1_HUMAN
|
||||||
| NC score | 0.029358 (rank : 104) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36915 | Gene names | GNL1, HSR1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein HSR1). | |||||
|
AN32A_MOUSE
|
||||||
| NC score | 0.028493 (rank : 105) | θ value | 1.06291 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| NC score | 0.028493 (rank : 106) | θ value | 1.06291 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
AN32E_HUMAN
|
||||||
| NC score | 0.028267 (rank : 107) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.027236 (rank : 108) | θ value | 1.38821 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
ENW1_HUMAN
|
||||||
| NC score | 0.025836 (rank : 109) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQF0, O95244, O95245, Q8NHY7, Q9NRZ2, Q9NZG3 | Gene names | ERVWE1 | |||
|
Domain Architecture |

|
|||||
| Description | HERV-W_7q21.2 provirus ancestral Env polyprotein precursor (Envelope polyprotein gPr73) (HERV-7q Envelope protein) (HERV-W envelope protein) (Syncytin) (Syncytin-1) (Enverin) (Env-W) [Contains: Surface protein (SU) (gp50); Transmembrane protein (TM) (gp24)]. | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.023961 (rank : 110) | θ value | 0.62314 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
ISY1_MOUSE
|
||||||
| NC score | 0.021316 (rank : 111) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q69ZQ2, Q8VDX4, Q9D3E2 | Gene names | Kiaa1160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor ISY1 homolog. | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.021299 (rank : 112) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
ISY1_HUMAN
|
||||||
| NC score | 0.020996 (rank : 113) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULR0, Q96IL2, Q9BT05 | Gene names | KIAA1160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor ISY1 homolog. | |||||
|
PPM1F_HUMAN
|
||||||
| NC score | 0.018336 (rank : 114) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
TICN2_HUMAN
|
||||||
| NC score | 0.018253 (rank : 115) | θ value | 1.06291 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TBL2_HUMAN
|
||||||
| NC score | 0.017218 (rank : 116) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4P3, Q9UQE2 | Gene names | TBL2, WBSCR13 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin beta-like 2 protein (WS beta-transducin repeats protein) (WS-betaTRP) (Williams-Beuren syndrome chromosome region 13 protein). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.016216 (rank : 117) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.015132 (rank : 118) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.014926 (rank : 119) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.014220 (rank : 120) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
MRC2_HUMAN
|
||||||
| NC score | 0.012501 (rank : 121) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.012272 (rank : 122) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
UBE2W_MOUSE
|
||||||
| NC score | 0.011801 (rank : 123) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDW4, Q9D5H3 | Gene names | Ube2w | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin-conjugating enzyme E2 W (EC 6.3.2.19) (Ubiquitin- protein ligase W) (Ubiquitin carrier protein W). | |||||
|
MRE11_HUMAN
|
||||||
| NC score | 0.011543 (rank : 124) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49959, O43475 | Gene names | MRE11A, HNGS1, MRE11 | |||
|
Domain Architecture |

|
|||||
| Description | Double-strand break repair protein MRE11A (MRE11 homolog 1) (MRE11 meiotic recombination 11 homolog A). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.011537 (rank : 125) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.011104 (rank : 126) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
UBE2W_HUMAN
|
||||||
| NC score | 0.010727 (rank : 127) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96B02, Q9H823, Q9HAG6, Q9NV07 | Gene names | UBE2W | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin-conjugating enzyme E2 W (EC 6.3.2.19) (Ubiquitin- protein ligase W) (Ubiquitin carrier protein W). | |||||
|
MRC2_MOUSE
|
||||||
| NC score | 0.010305 (rank : 128) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
ADA19_HUMAN
|
||||||
| NC score | 0.009490 (rank : 129) | θ value | 1.38821 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
TSP4_MOUSE
|
||||||
| NC score | 0.009246 (rank : 130) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1T2, Q9QYS3, Q9WUE0 | Gene names | Thbs4, Tsp4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
TSP4_HUMAN
|
||||||
| NC score | 0.009235 (rank : 131) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35443, Q86TG2 | Gene names | THBS4, TSP4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
PARP9_MOUSE
|
||||||
| NC score | 0.008045 (rank : 132) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
APBB1_HUMAN
|
||||||
| NC score | 0.006762 (rank : 133) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
PDIA4_MOUSE
|
||||||
| NC score | 0.006464 (rank : 134) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08003, P15841 | Gene names | Pdia4, Cai, Erp72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
DUS3_HUMAN
|
||||||
| NC score | 0.005572 (rank : 135) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
KLDC5_MOUSE
|
||||||
| NC score | 0.005307 (rank : 136) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFQ9, Q6ZPT4 | Gene names | Klhdc5, Kiaa1340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 5. | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.004803 (rank : 137) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
CLCN5_MOUSE
|
||||||
| NC score | 0.004428 (rank : 138) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVD4 | Gene names | Clcn5, Clc5 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 5 (ClC-5). | |||||
|
PRELP_MOUSE
|
||||||
| NC score | 0.002554 (rank : 139) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JK53 | Gene names | Prelp | |||
|
Domain Architecture |

|
|||||
| Description | Prolargin precursor (Proline-arginine-rich end leucine-rich repeat protein). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.002113 (rank : 140) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
SUHW3_MOUSE
|
||||||
| NC score | 0.001915 (rank : 141) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.001363 (rank : 142) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PTPRD_MOUSE
|
||||||
| NC score | 0.000888 (rank : 143) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.000870 (rank : 144) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.000395 (rank : 145) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ZBTB1_MOUSE
|
||||||
| NC score | -0.000713 (rank : 146) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VL9, Q8CDP7, Q99LD2 | Gene names | Zbtb1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 1. | |||||
|
M3K6_HUMAN
|
||||||
| NC score | -0.003421 (rank : 147) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95382 | Gene names | MAP3K6, MAPKKK6 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 6 (EC 2.7.11.25). | |||||