Please be patient as the page loads
|
BCL7A_MOUSE
|
||||||
| SwissProt Accessions | Q9CXE2, Q8C361, Q8C8M8, Q8VD15 | Gene names | Bcl7a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BCL7A_MOUSE
|
||||||
| θ value | 5.30912e-93 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9CXE2, Q8C361, Q8C8M8, Q8VD15 | Gene names | Bcl7a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
BCL7A_HUMAN
|
||||||
| θ value | 3.0147e-88 (rank : 2) | NC score | 0.982930 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
BCL7B_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 3) | NC score | 0.896972 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQE9, O43769, Q13845, Q6ZW75 | Gene names | BCL7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
BCL7B_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 4) | NC score | 0.893378 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921K9, O89022, Q3TV31, Q3U2W0 | Gene names | Bcl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
BCL7C_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 5) | NC score | 0.859763 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUZ0, O43770, Q6PD89 | Gene names | BCL7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C. | |||||
|
BCL7C_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 6) | NC score | 0.812666 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.073484 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.106074 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.037563 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.059363 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.020456 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.056918 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.026987 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.019976 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.057365 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.040118 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.029668 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.014092 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.019825 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.046629 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.031706 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
FBX7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.064921 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.046363 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.030907 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.031125 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.011455 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.019502 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.018487 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DUET_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.001674 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
HBS1L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.017514 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.053878 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.041544 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.011996 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.028502 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.015669 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
FOXP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.020568 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
FOXP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.019862 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
RBL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.019006 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64700 | Gene names | Rbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.019450 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ZN648_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.001457 (rank : 75) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T619 | Gene names | ZNF648 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 648. | |||||
|
ANKY2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.014941 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
DJC12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.028645 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |
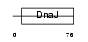
|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
EXO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.034799 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
FOXN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.015324 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q499D0, Q8C5N8, Q9CU83 | Gene names | Ches1, Foxn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.014350 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.016511 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.007819 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.049145 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.047966 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.027949 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
DCR1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.027053 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.042083 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DTBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.018362 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WZ8, Q80ZN4 | Gene names | Dtnbp1 | |||
|
Domain Architecture |
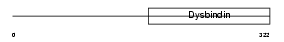
|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein homolog). | |||||
|
FOXR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.015258 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UTB7 | Gene names | Foxr1, Foxn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein R1 (Forkhead box protein N5). | |||||
|
PHLPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.006602 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.035226 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
BCAS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.023086 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
BCL6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.002049 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |
|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.012006 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.047196 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.034842 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
IF39_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.027286 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.006859 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.027746 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.025521 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.039860 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
JAK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | -0.000937 (rank : 76) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52332, Q62126 | Gene names | Jak1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK1 (EC 2.7.10.2) (Janus kinase 1) (JAK-1). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.012686 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MYSM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.023391 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
NHS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.020371 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.020236 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
TMUB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.013299 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V209, Q8K0X0, Q91YQ0, Q9D8C5 | Gene names | Tmub2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
ZEP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.005544 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
ZHX2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.008398 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6X8 | Gene names | ZHX2, AFR1, KIAA0854, RAF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.052776 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PRPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.050070 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
BCL7A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.30912e-93 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9CXE2, Q8C361, Q8C8M8, Q8VD15 | Gene names | Bcl7a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
BCL7A_HUMAN
|
||||||
| NC score | 0.982930 (rank : 2) | θ value | 3.0147e-88 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
BCL7B_HUMAN
|
||||||
| NC score | 0.896972 (rank : 3) | θ value | 5.22648e-32 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQE9, O43769, Q13845, Q6ZW75 | Gene names | BCL7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
BCL7B_MOUSE
|
||||||
| NC score | 0.893378 (rank : 4) | θ value | 5.22648e-32 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921K9, O89022, Q3TV31, Q3U2W0 | Gene names | Bcl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
BCL7C_HUMAN
|
||||||
| NC score | 0.859763 (rank : 5) | θ value | 2.12685e-25 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUZ0, O43770, Q6PD89 | Gene names | BCL7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C. | |||||
|
BCL7C_MOUSE
|
||||||
| NC score | 0.812666 (rank : 6) | θ value | 1.05554e-24 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.106074 (rank : 7) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.073484 (rank : 8) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
FBX7_HUMAN
|
||||||
| NC score | 0.064921 (rank : 9) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.059363 (rank : 10) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.057365 (rank : 11) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.056918 (rank : 12) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.053878 (rank : 13) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.052776 (rank : 14) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.050070 (rank : 15) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.049145 (rank : 16) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.047966 (rank : 17) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.047196 (rank : 18) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.046629 (rank : 19) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.046363 (rank : 20) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.042083 (rank : 21) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.041544 (rank : 22) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.040118 (rank : 23) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.039860 (rank : 24) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.037563 (rank : 25) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.035226 (rank : 26) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.034842 (rank : 27) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
EXO1_HUMAN
|
||||||
| NC score | 0.034799 (rank : 28) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.031706 (rank : 29) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.031125 (rank : 30) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.030907 (rank : 31) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.029668 (rank : 32) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
DJC12_MOUSE
|
||||||
| NC score | 0.028645 (rank : 33) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.028502 (rank : 34) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.027949 (rank : 35) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.027746 (rank : 36) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.027286 (rank : 37) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
DCR1A_MOUSE
|
||||||
| NC score | 0.027053 (rank : 38) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.026987 (rank : 39) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.025521 (rank : 40) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
MYSM1_MOUSE
|
||||||
| NC score | 0.023391 (rank : 41) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
BCAS1_HUMAN
|
||||||
| NC score | 0.023086 (rank : 42) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
FOXP4_HUMAN
|
||||||
| NC score | 0.020568 (rank : 43) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.020456 (rank : 44) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
NHS_HUMAN
|
||||||
| NC score | 0.020371 (rank : 45) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.020236 (rank : 46) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.019976 (rank : 47) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
FOXP4_MOUSE
|
||||||
| NC score | 0.019862 (rank : 48) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.019825 (rank : 49) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.019502 (rank : 50) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.019450 (rank : 51) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
RBL2_MOUSE
|
||||||
| NC score | 0.019006 (rank : 52) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64700 | Gene names | Rbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.018487 (rank : 53) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DTBP1_MOUSE
|
||||||
| NC score | 0.018362 (rank : 54) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WZ8, Q80ZN4 | Gene names | Dtnbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein homolog). | |||||
|
HBS1L_HUMAN
|
||||||
| NC score | 0.017514 (rank : 55) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.016511 (rank : 56) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.015669 (rank : 57) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
FOXN3_MOUSE
|
||||||
| NC score | 0.015324 (rank : 58) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q499D0, Q8C5N8, Q9CU83 | Gene names | Ches1, Foxn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
FOXR1_MOUSE
|
||||||
| NC score | 0.015258 (rank : 59) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UTB7 | Gene names | Foxr1, Foxn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein R1 (Forkhead box protein N5). | |||||
|
ANKY2_MOUSE
|
||||||
| NC score | 0.014941 (rank : 60) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.014350 (rank : 61) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.014092 (rank : 62) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
TMUB2_MOUSE
|
||||||
| NC score | 0.013299 (rank : 63) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V209, Q8K0X0, Q91YQ0, Q9D8C5 | Gene names | Tmub2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.012686 (rank : 64) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.012006 (rank : 65) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.011996 (rank : 66) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.011455 (rank : 67) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
ZHX2_HUMAN
|
||||||
| NC score | 0.008398 (rank : 68) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6X8 | Gene names | ZHX2, AFR1, KIAA0854, RAF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.007819 (rank : 69) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.006859 (rank : 70) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
PHLPL_MOUSE
|
||||||
| NC score | 0.006602 (rank : 71) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
ZEP2_HUMAN
|
||||||
| NC score | 0.005544 (rank : 72) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
BCL6_MOUSE
|
||||||
| NC score | 0.002049 (rank : 73) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||
|
DUET_HUMAN
|
||||||
| NC score | 0.001674 (rank : 74) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
ZN648_HUMAN
|
||||||
| NC score | 0.001457 (rank : 75) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T619 | Gene names | ZNF648 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 648. | |||||
|
JAK1_MOUSE
|
||||||
| NC score | -0.000937 (rank : 76) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52332, Q62126 | Gene names | Jak1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK1 (EC 2.7.10.2) (Janus kinase 1) (JAK-1). | |||||