Please be patient as the page loads
|
ERCC5_HUMAN
|
||||||
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ERCC5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 162 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.922044 (rank : 2) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
FEN1_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 3) | NC score | 0.503022 (rank : 3) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39749 | Gene names | Fen1, Fen-1 | |||
|
Domain Architecture |
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1). | |||||
|
FEN1_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 4) | NC score | 0.491835 (rank : 4) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39748 | Gene names | FEN1 | |||
|
Domain Architecture |
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1) (Maturation factor 1) (MF1) (hFEN-1) (DNase IV). | |||||
|
EXO1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 5) | NC score | 0.275711 (rank : 5) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
EXO1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 6) | NC score | 0.267943 (rank : 6) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 7) | NC score | 0.072227 (rank : 23) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 8) | NC score | 0.020302 (rank : 150) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.052763 (rank : 58) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.061491 (rank : 36) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.088035 (rank : 10) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.076594 (rank : 17) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.085954 (rank : 12) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.073237 (rank : 21) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
CE005_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.068997 (rank : 28) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.092277 (rank : 8) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.033512 (rank : 125) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.080489 (rank : 15) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.069858 (rank : 26) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.083948 (rank : 14) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.055741 (rank : 48) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.050426 (rank : 67) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.088840 (rank : 9) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.054133 (rank : 51) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.053769 (rank : 53) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.043190 (rank : 92) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
CE005_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.059249 (rank : 40) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.073436 (rank : 20) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.126693 (rank : 7) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.034080 (rank : 123) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.065903 (rank : 32) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NOL8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.048450 (rank : 73) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.052884 (rank : 57) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
LMAN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.059629 (rank : 39) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.053092 (rank : 55) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
CABYR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.052670 (rank : 59) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
CM003_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.047570 (rank : 77) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C263 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3 homolog. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.050501 (rank : 66) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.009861 (rank : 172) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.070707 (rank : 25) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.053649 (rank : 54) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
LMAN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.050901 (rank : 64) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.034058 (rank : 124) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.073595 (rank : 19) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.023706 (rank : 142) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TSNAX_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.040924 (rank : 103) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZE7, Q3UJR2 | Gene names | Tsnax, Trax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translin-associated protein X (Translin-associated factor X). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.053993 (rank : 52) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
K0494_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.051415 (rank : 62) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BGQ6, Q3TQN6, Q8BJV7, Q8C0R1 | Gene names | Kiaa0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
KI13A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.022983 (rank : 145) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.034240 (rank : 122) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.027638 (rank : 134) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.086478 (rank : 11) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.084584 (rank : 13) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.064081 (rank : 33) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.045667 (rank : 83) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.043921 (rank : 90) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.062229 (rank : 35) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.041387 (rank : 101) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.040577 (rank : 105) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.014236 (rank : 166) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
BFSP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.040349 (rank : 107) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
EDEM3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.024600 (rank : 138) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.043018 (rank : 93) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
FAM9A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.045510 (rank : 84) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.056801 (rank : 43) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.066452 (rank : 31) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.041794 (rank : 100) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.039493 (rank : 113) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.028914 (rank : 132) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.049090 (rank : 71) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
SLC2B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.042915 (rank : 94) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
TPH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.016914 (rank : 156) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17752, O95188, O95189, Q16736 | Gene names | TPH1, TPH, TRPH | |||
|
Domain Architecture |
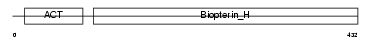
|
|||||
| Description | Tryptophan 5-hydroxylase 1 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 1). | |||||
|
TRIM6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.014380 (rank : 165) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9C030, Q86WZ8, Q9HCR1 | Gene names | TRIM6, RNF89 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 6 (RING finger protein 89). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.050130 (rank : 68) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.047383 (rank : 79) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.015372 (rank : 163) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
DX26B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.045393 (rank : 85) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JSJ4, Q5CZA2, Q6IPS3, Q6ZTU5, Q6ZWE4 | Gene names | DDX26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.074159 (rank : 18) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
GBP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.027657 (rank : 133) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.062380 (rank : 34) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LRC50_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.022182 (rank : 146) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEP3, Q69YI8, Q69YJ0, Q96LP3 | Gene names | LRRC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.042393 (rank : 98) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PTPRG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.011752 (rank : 170) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.022067 (rank : 147) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
SVIL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.019611 (rank : 151) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.044080 (rank : 89) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.025238 (rank : 136) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.024901 (rank : 137) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.034337 (rank : 120) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
CENPB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.048928 (rank : 72) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CIP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.034947 (rank : 118) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
CXA8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.008093 (rank : 176) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48165, Q9NP25 | Gene names | GJA8 | |||
|
Domain Architecture |
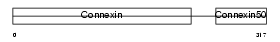
|
|||||
| Description | Gap junction alpha-8 protein (Connexin-50) (Cx50) (Lens fiber protein MP70). | |||||
|
DC1I2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.015485 (rank : 162) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88487, Q3TGH7 | Gene names | Dync1i2, Dnci2, Dncic2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 2 (Dynein intermediate chain 2, cytosolic) (DH IC-2) (Cytoplasmic dynein intermediate chain 2). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.008093 (rank : 175) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.047746 (rank : 76) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
K0406_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.021632 (rank : 148) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V83, Q9CSY0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0406 homolog. | |||||
|
KINH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.041171 (rank : 102) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.044497 (rank : 88) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.044724 (rank : 87) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.047500 (rank : 78) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.023757 (rank : 141) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
SEMG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.033334 (rank : 127) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.016314 (rank : 158) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ZHX2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.015529 (rank : 161) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
ABCAC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.009439 (rank : 173) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UK0, Q8IZW6, Q96JT3, Q9Y4M5 | Gene names | ABCA12, ABC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 12 (ATP-binding cassette transporter 12) (ATP-binding cassette 12). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.045152 (rank : 86) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.045784 (rank : 82) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.067897 (rank : 30) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
HAP28_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.033510 (rank : 126) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3UHX2, Q3KQJ5 | Gene names | Pdap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28 kDa heat- and acid-stable phosphoprotein (PDGF-associated protein) (PAP) (PDGFA-associated protein 1) (PAP1). | |||||
|
LZTS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.042202 (rank : 99) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.013919 (rank : 167) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.040173 (rank : 109) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.048286 (rank : 74) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
POLK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.034414 (rank : 119) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UBT6, Q86VJ8, Q8IZY0, Q8IZY1, Q8NB30, Q96L01, Q96Q86, Q96Q87, Q9UHC5 | Gene names | POLK, DINB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
PPIP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.042514 (rank : 97) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99M15, Q9Z189 | Gene names | Pstpip2, Mayp | |||
|
Domain Architecture |
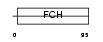
|
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 2 (Macrophage actin-associated tyrosine-phosphorylated protein) (pp37). | |||||
|
PRP8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.040491 (rank : 106) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P2Q9, O14547, O75965 | Gene names | PRPF8, PRPC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-processing-splicing factor 8 (Splicing factor Prp8) (PRP8 homolog) (220 kDa U5 snRNP-specific protein) (p220). | |||||
|
PRP8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.040614 (rank : 104) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PV0, Q5ND30, Q5ND31, Q7TQK2, Q8BRZ5, Q8K001 | Gene names | Prpf8, Prp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-processing-splicing factor 8 (Splicing factor Prp8). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.039202 (rank : 114) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.048019 (rank : 75) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.042733 (rank : 95) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
ZFY2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | -0.000323 (rank : 178) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
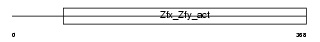
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.023860 (rank : 140) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.040231 (rank : 108) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
ASPH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.043520 (rank : 91) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.056559 (rank : 44) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.049907 (rank : 70) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.018506 (rank : 153) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.056503 (rank : 45) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
ICA69_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.013448 (rank : 168) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97411, P97362, P97954, Q99M40, Q9R2C4 | Gene names | Ica1, Icap69 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Islet cell autoantigen 1 (69 kDa islet cell autoantigen) (ICA69) (p69) (Islet cell autoantigen p69) (ICAp69). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.023308 (rank : 144) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.040126 (rank : 110) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KIF23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.036264 (rank : 115) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.047282 (rank : 80) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.035508 (rank : 117) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.025739 (rank : 135) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.029953 (rank : 131) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NFL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.031516 (rank : 130) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NOC2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.018602 (rank : 152) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.023491 (rank : 143) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.050036 (rank : 69) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.021168 (rank : 149) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.016604 (rank : 157) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.039655 (rank : 111) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.015924 (rank : 160) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.042556 (rank : 96) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
DOCK8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.007401 (rank : 177) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
HSH2D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.010929 (rank : 171) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.008378 (rank : 174) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.034270 (rank : 121) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.032647 (rank : 128) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.079821 (rank : 16) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NPHP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.017811 (rank : 154) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.046836 (rank : 81) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PHAR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.015289 (rank : 164) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9C0D0, Q3MJ93, Q5JSJ2 | Gene names | PHACTR1, KIAA1733, RPEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatase and actin regulator 1. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.035631 (rank : 116) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SETX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.016107 (rank : 159) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
SH3BG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.017302 (rank : 155) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.039499 (rank : 112) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TCAL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.024579 (rank : 139) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TRIM6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.012336 (rank : 169) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BGE7, Q99PQ6 | Gene names | Trim6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 6. | |||||
|
UBXD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.031701 (rank : 129) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96LJ8 | Gene names | UBXD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 3. | |||||
|
ZNF18_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | -0.001762 (rank : 179) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810A1, Q9D972 | Gene names | Znf18, Zfp535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein 535). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.071921 (rank : 24) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.051601 (rank : 60) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.060131 (rank : 38) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.058739 (rank : 41) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.055519 (rank : 49) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.057708 (rank : 42) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.073191 (rank : 22) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.056235 (rank : 46) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.055344 (rank : 50) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.053022 (rank : 56) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.068743 (rank : 29) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.051308 (rank : 63) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.056232 (rank : 47) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.050863 (rank : 65) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.060882 (rank : 37) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.051563 (rank : 61) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.069418 (rank : 27) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 162 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.922044 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
FEN1_MOUSE
|
||||||
| NC score | 0.503022 (rank : 3) | θ value | 1.42992e-13 (rank : 3) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39749 | Gene names | Fen1, Fen-1 | |||
|
Domain Architecture |
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1). | |||||
|
FEN1_HUMAN
|
||||||
| NC score | 0.491835 (rank : 4) | θ value | 9.26847e-13 (rank : 4) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39748 | Gene names | FEN1 | |||
|
Domain Architecture |
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1) (Maturation factor 1) (MF1) (hFEN-1) (DNase IV). | |||||
|
EXO1_MOUSE
|
||||||
| NC score | 0.275711 (rank : 5) | θ value | 9.29e-05 (rank : 5) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
EXO1_HUMAN
|
||||||
| NC score | 0.267943 (rank : 6) | θ value | 0.000270298 (rank : 6) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.126693 (rank : 7) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.092277 (rank : 8) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.088840 (rank : 9) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.088035 (rank : 10) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.086478 (rank : 11) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.085954 (rank : 12) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.084584 (rank : 13) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.083948 (rank : 14) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.080489 (rank : 15) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.079821 (rank : 16) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.076594 (rank : 17) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.074159 (rank : 18) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.073595 (rank : 19) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.073436 (rank : 20) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.073237 (rank : 21) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.073191 (rank : 22) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.072227 (rank : 23) | θ value | 0.00175202 (rank : 7) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.071921 (rank : 24) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.070707 (rank : 25) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.069858 (rank : 26) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.069418 (rank : 27) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.068997 (rank : 28) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.068743 (rank : 29) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.067897 (rank : 30) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.066452 (rank : 31) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.065903 (rank : 32) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.064081 (rank : 33) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.062380 (rank : 34) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.062229 (rank : 35) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.061491 (rank : 36) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.060882 (rank : 37) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.060131 (rank : 38) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
LMAN1_HUMAN
|
||||||
| NC score | 0.059629 (rank : 39) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.059249 (rank : 40) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.058739 (rank : 41) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.057708 (rank : 42) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.056801 (rank : 43) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.056559 (rank : 44) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.056503 (rank : 45) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.056235 (rank : 46) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.056232 (rank : 47) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.055741 (rank : 48) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.055519 (rank : 49) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.055344 (rank : 50) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.054133 (rank : 51) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.053993 (rank : 52) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.053769 (rank : 53) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.053649 (rank : 54) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.053092 (rank : 55) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.053022 (rank : 56) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.052884 (rank : 57) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.052763 (rank : 58) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
CABYR_HUMAN
|
||||||
| NC score | 0.052670 (rank : 59) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.051601 (rank : 60) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.051563 (rank : 61) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
K0494_MOUSE
|
||||||
| NC score | 0.051415 (rank : 62) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BGQ6, Q3TQN6, Q8BJV7, Q8C0R1 | Gene names | Kiaa0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.051308 (rank : 63) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
LMAN1_MOUSE
|
||||||
| NC score | 0.050901 (rank : 64) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.050863 (rank : 65) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.050501 (rank : 66) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.050426 (rank : 67) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.050130 (rank : 68) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.050036 (rank : 69) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.049907 (rank : 70) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.049090 (rank : 71) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
CENPB_HUMAN
|
||||||
| NC score | 0.048928 (rank : 72) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
NOL8_MOUSE
|
||||||
| NC score | 0.048450 (rank : 73) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.048286 (rank : 74) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.048019 (rank : 75) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.047746 (rank : 76) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
CM003_MOUSE
|
||||||
| NC score | 0.047570 (rank : 77) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C263 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3 homolog. | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.047500 (rank : 78) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.047383 (rank : 79) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.047282 (rank : 80) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.046836 (rank : 81) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.045784 (rank : 82) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.045667 (rank : 83) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
FAM9A_HUMAN
|
||||||
| NC score | 0.045510 (rank : 84) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
DX26B_HUMAN
|
||||||
| NC score | 0.045393 (rank : 85) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JSJ4, Q5CZA2, Q6IPS3, Q6ZTU5, Q6ZWE4 | Gene names | DDX26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.045152 (rank : 86) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.044724 (rank : 87) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.044497 (rank : 88) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.044080 (rank : 89) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.043921 (rank : 90) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
ASPH_HUMAN
|
||||||
| NC score | 0.043520 (rank : 91) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.043190 (rank : 92) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.043018 (rank : 93) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
SLC2B_HUMAN
|
||||||
| NC score | 0.042915 (rank : 94) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.042733 (rank : 95) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.042556 (rank : 96) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
PPIP2_MOUSE
|
||||||
| NC score | 0.042514 (rank : 97) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99M15, Q9Z189 | Gene names | Pstpip2, Mayp | |||
|
Domain Architecture |

|
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 2 (Macrophage actin-associated tyrosine-phosphorylated protein) (pp37). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.042393 (rank : 98) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
LZTS1_HUMAN
|
||||||
| NC score | 0.042202 (rank : 99) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.041794 (rank : 100) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.041387 (rank : 101) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.041171 (rank : 102) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
TSNAX_MOUSE
|
||||||
| NC score | 0.040924 (rank : 103) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZE7, Q3UJR2 | Gene names | Tsnax, Trax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translin-associated protein X (Translin-associated factor X). | |||||
|
PRP8_MOUSE
|
||||||
| NC score | 0.040614 (rank : 104) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PV0, Q5ND30, Q5ND31, Q7TQK2, Q8BRZ5, Q8K001 | Gene names | Prpf8, Prp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-processing-splicing factor 8 (Splicing factor Prp8). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.040577 (rank : 105) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PRP8_HUMAN
|
||||||
| NC score | 0.040491 (rank : 106) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P2Q9, O14547, O75965 | Gene names | PRPF8, PRPC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-processing-splicing factor 8 (Splicing factor Prp8) (PRP8 homolog) (220 kDa U5 snRNP-specific protein) (p220). | |||||
|
BFSP2_MOUSE
|
||||||
| NC score | 0.040349 (rank : 107) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.040231 (rank : 108) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.040173 (rank : 109) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.040126 (rank : 110) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.039655 (rank : 111) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.039499 (rank : 112) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.039493 (rank : 113) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.039202 (rank : 114) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
KIF23_HUMAN
|
||||||
| NC score | 0.036264 (rank : 115) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.035631 (rank : 116) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.035508 (rank : 117) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
CIP4_HUMAN
|
||||||
| NC score | 0.034947 (rank : 118) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
POLK_HUMAN
|
||||||
| NC score | 0.034414 (rank : 119) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UBT6, Q86VJ8, Q8IZY0, Q8IZY1, Q8NB30, Q96L01, Q96Q86, Q96Q87, Q9UHC5 | Gene names | POLK, DINB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.034337 (rank : 120) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.034270 (rank : 121) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.034240 (rank : 122) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.034080 (rank : 123) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.034058 (rank : 124) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.033512 (rank : 125) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
HAP28_MOUSE
|
||||||
| NC score | 0.033510 (rank : 126) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3UHX2, Q3KQJ5 | Gene names | Pdap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28 kDa heat- and acid-stable phosphoprotein (PDGF-associated protein) (PAP) (PDGFA-associated protein 1) (PAP1). | |||||
|
SEMG1_HUMAN
|
||||||
| NC score | 0.033334 (rank : 127) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.032647 (rank : 128) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
UBXD3_HUMAN
|
||||||
| NC score | 0.031701 (rank : 129) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96LJ8 | Gene names | UBXD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 3. | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.031516 (rank : 130) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.029953 (rank : 131) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.028914 (rank : 132) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GBP2_HUMAN
|
||||||
| NC score | 0.027657 (rank : 133) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.027638 (rank : 134) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.025739 (rank : 135) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.025238 (rank : 136) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.024901 (rank : 137) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
EDEM3_HUMAN
|
||||||
| NC score | 0.024600 (rank : 138) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
TCAL3_MOUSE
|
||||||
| NC score | 0.024579 (rank : 139) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.023860 (rank : 140) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.023757 (rank : 141) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.023706 (rank : 142) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.023491 (rank : 143) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.023308 (rank : 144) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
KI13A_HUMAN
|
||||||
| NC score | 0.022983 (rank : 145) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
LRC50_HUMAN
|
||||||
| NC score | 0.022182 (rank : 146) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEP3, Q69YI8, Q69YJ0, Q96LP3 | Gene names | LRRC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.022067 (rank : 147) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
K0406_MOUSE
|
||||||
| NC score | 0.021632 (rank : 148) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V83, Q9CSY0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0406 homolog. | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.021168 (rank : 149) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.020302 (rank : 150) | θ value | 0.00175202 (rank : 8) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
SVIL_HUMAN
|
||||||
| NC score | 0.019611 (rank : 151) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
NOC2L_HUMAN
|
||||||
| NC score | 0.018602 (rank : 152) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.018506 (rank : 153) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
NPHP3_MOUSE
|
||||||
| NC score | 0.017811 (rank : 154) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
SH3BG_MOUSE
|
||||||
| NC score | 0.017302 (rank : 155) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
TPH1_HUMAN
|
||||||
| NC score | 0.016914 (rank : 156) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17752, O95188, O95189, Q16736 | Gene names | TPH1, TPH, TRPH | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophan 5-hydroxylase 1 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 1). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.016604 (rank : 157) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.016314 (rank : 158) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.016107 (rank : 159) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.015924 (rank : 160) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
ZHX2_MOUSE
|
||||||
| NC score | 0.015529 (rank : 161) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
DC1I2_MOUSE
|
||||||
| NC score | 0.015485 (rank : 162) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88487, Q3TGH7 | Gene names | Dync1i2, Dnci2, Dncic2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 2 (Dynein intermediate chain 2, cytosolic) (DH IC-2) (Cytoplasmic dynein intermediate chain 2). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.015372 (rank : 163) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
PHAR1_HUMAN
|
||||||
| NC score | 0.015289 (rank : 164) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9C0D0, Q3MJ93, Q5JSJ2 | Gene names | PHACTR1, KIAA1733, RPEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatase and actin regulator 1. | |||||
|
TRIM6_HUMAN
|
||||||
| NC score | 0.014380 (rank : 165) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9C030, Q86WZ8, Q9HCR1 | Gene names | TRIM6, RNF89 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 6 (RING finger protein 89). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.014236 (rank : 166) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.013919 (rank : 167) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
ICA69_MOUSE
|
||||||
| NC score | 0.013448 (rank : 168) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97411, P97362, P97954, Q99M40, Q9R2C4 | Gene names | Ica1, Icap69 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Islet cell autoantigen 1 (69 kDa islet cell autoantigen) (ICA69) (p69) (Islet cell autoantigen p69) (ICAp69). | |||||
|
TRIM6_MOUSE
|
||||||
| NC score | 0.012336 (rank : 169) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BGE7, Q99PQ6 | Gene names | Trim6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 6. | |||||
|
PTPRG_MOUSE
|
||||||
| NC score | 0.011752 (rank : 170) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
HSH2D_MOUSE
|
||||||
| NC score | 0.010929 (rank : 171) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.009861 (rank : 172) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
ABCAC_HUMAN
|
||||||
| NC score | 0.009439 (rank : 173) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UK0, Q8IZW6, Q96JT3, Q9Y4M5 | Gene names | ABCA12, ABC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 12 (ATP-binding cassette transporter 12) (ATP-binding cassette 12). | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.008378 (rank : 174) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.008093 (rank : 175) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
CXA8_HUMAN
|
||||||
| NC score | 0.008093 (rank : 176) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48165, Q9NP25 | Gene names | GJA8 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-8 protein (Connexin-50) (Cx50) (Lens fiber protein MP70). | |||||
|
DOCK8_HUMAN
|
||||||
| NC score | 0.007401 (rank : 177) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
ZFY2_MOUSE
|
||||||
| NC score | -0.000323 (rank : 178) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
ZNF18_MOUSE
|
||||||
| NC score | -0.001762 (rank : 179) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810A1, Q9D972 | Gene names | Znf18, Zfp535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein 535). | |||||