Please be patient as the page loads
|
LMAN1_HUMAN
|
||||||
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LMAN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
LMAN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.988946 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
LMA1L_HUMAN
|
||||||
| θ value | 7.97034e-73 (rank : 3) | NC score | 0.920505 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HAT1, Q6UWN2 | Gene names | LMAN1L, ERGL | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMA1L_MOUSE
|
||||||
| θ value | 1.31987e-59 (rank : 4) | NC score | 0.923081 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VCD3 | Gene names | Lman1l, Ergl | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMAN2_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 5) | NC score | 0.850177 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBH5, Q9CXG7 | Gene names | Lman2 | |||
|
Domain Architecture |
|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (Lectin, mannose- binding 2). | |||||
|
LMAN2_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 6) | NC score | 0.849296 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12907, Q53HH1 | Gene names | LMAN2, C5orf8 | |||
|
Domain Architecture |
|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (GP36b glycoprotein) (Lectin, mannose-binding 2). | |||||
|
LMA2L_HUMAN
|
||||||
| θ value | 4.13158e-37 (rank : 7) | NC score | 0.838126 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0V9, Q53GV3, Q53S67, Q63HN6, Q8NBQ6, Q9BQ14 | Gene names | LMAN2L, VIPL | |||
|
Domain Architecture |
|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMA2L_MOUSE
|
||||||
| θ value | 1.01765e-35 (rank : 8) | NC score | 0.836992 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59481, Q3TZ91 | Gene names | Lman2l, Vipl | |||
|
Domain Architecture |
|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.077769 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.076390 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.076476 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.068116 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.073222 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.060112 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.050983 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.079958 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
LAMA4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.041191 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.031360 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.071564 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
TUFT1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.072148 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.067250 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
CF182_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.063045 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
ERCC5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.059629 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
KIF23_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.056378 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
MAF_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.040532 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.057629 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.048693 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.054962 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
RILP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.042362 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96NA2, Q71RE6, Q9BSL3, Q9BYS3 | Gene names | RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab-interacting lysosomal protein. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.019522 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.064881 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.064955 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.066306 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MAF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.038258 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.057016 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.056325 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.018100 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.017936 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.053902 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
AATC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.042608 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17174 | Gene names | GOT1 | |||
|
Domain Architecture |
|
|||||
| Description | Aspartate aminotransferase, cytoplasmic (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 1). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.059815 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.062029 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.041781 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.041622 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.055954 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.025490 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.048630 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CAB45_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.021154 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRK5, Q8NBQ3, Q9NZP7 | Gene names | SDF4, CAB45 | |||
|
Domain Architecture |
|
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.051219 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.060222 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
LC7L2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.046724 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
LC7L2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.046877 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.033204 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.045237 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.054651 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RAB3I_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.039889 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.024811 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.046816 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
AMRP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.046893 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55302 | Gene names | Lrpap1 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2-macroglobulin receptor-associated protein precursor (Alpha-2- MRAP) (Low density lipoprotein receptor-related protein-associated protein 1) (RAP) (Heparin-binding protein 44) (HBP-44). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.047874 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.054907 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.048925 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.023713 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.052072 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FER_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.006382 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P16591 | Gene names | FER, TYK3 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FER (EC 2.7.10.2) (p94-FER) (c- FER). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.055663 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.008999 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.048465 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.052263 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.053428 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.053342 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.034026 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.048520 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.048580 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.019395 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.043936 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
AATC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.032461 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05201 | Gene names | Got1 | |||
|
Domain Architecture |
|
|||||
| Description | Aspartate aminotransferase, cytoplasmic (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 1). | |||||
|
AMRP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.036966 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30533 | Gene names | LRPAP1, A2MRAP | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2-macroglobulin receptor-associated protein precursor (Alpha-2- MRAP) (Low density lipoprotein receptor-related protein-associated protein 1) (RAP). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.041449 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.038874 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.038708 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SEPT8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.012632 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92599, Q8IX36, Q8IX37, Q9BVB3 | Gene names | SEPT8, KIAA0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
SEPT8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.012078 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CHH9, Q80YC7, Q9ESF7 | Gene names | Sept8, Kiaa0202 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-8. | |||||
|
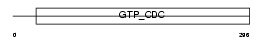
CCD39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.052079 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CE290_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.054040 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.051409 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CING_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.050102 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050365 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.053212 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
NIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.050593 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
LMAN1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
LMAN1_MOUSE
|
||||||
| NC score | 0.988946 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
LMA1L_MOUSE
|
||||||
| NC score | 0.923081 (rank : 3) | θ value | 1.31987e-59 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VCD3 | Gene names | Lman1l, Ergl | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMA1L_HUMAN
|
||||||
| NC score | 0.920505 (rank : 4) | θ value | 7.97034e-73 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HAT1, Q6UWN2 | Gene names | LMAN1L, ERGL | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMAN2_MOUSE
|
||||||
| NC score | 0.850177 (rank : 5) | θ value | 3.37985e-39 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBH5, Q9CXG7 | Gene names | Lman2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (Lectin, mannose- binding 2). | |||||
|
LMAN2_HUMAN
|
||||||
| NC score | 0.849296 (rank : 6) | θ value | 7.52953e-39 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12907, Q53HH1 | Gene names | LMAN2, C5orf8 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (GP36b glycoprotein) (Lectin, mannose-binding 2). | |||||
|
LMA2L_HUMAN
|
||||||
| NC score | 0.838126 (rank : 7) | θ value | 4.13158e-37 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0V9, Q53GV3, Q53S67, Q63HN6, Q8NBQ6, Q9BQ14 | Gene names | LMAN2L, VIPL | |||
|
Domain Architecture |

|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMA2L_MOUSE
|
||||||
| NC score | 0.836992 (rank : 8) | θ value | 1.01765e-35 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59481, Q3TZ91 | Gene names | Lman2l, Vipl | |||
|
Domain Architecture |

|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.079958 (rank : 9) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.077769 (rank : 10) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.076476 (rank : 11) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.076390 (rank : 12) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.073222 (rank : 13) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
TUFT1_MOUSE
|
||||||
| NC score | 0.072148 (rank : 14) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.071564 (rank : 15) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.068116 (rank : 16) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.067250 (rank : 17) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.066306 (rank : 18) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.064955 (rank : 19) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.064881 (rank : 20) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CF182_MOUSE
|
||||||
| NC score | 0.063045 (rank : 21) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.062029 (rank : 22) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.060222 (rank : 23) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.060112 (rank : 24) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.059815 (rank : 25) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.059629 (rank : 26) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.057629 (rank : 27) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.057016 (rank : 28) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
KIF23_HUMAN
|
||||||
| NC score | 0.056378 (rank : 29) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.056325 (rank : 30) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.055954 (rank : 31) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.055663 (rank : 32) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.054962 (rank : 33) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.054907 (rank : 34) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.054651 (rank : 35) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.054040 (rank : 36) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.053902 (rank : 37) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.053428 (rank : 38) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.053342 (rank : 39) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.053212 (rank : 40) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.052263 (rank : 41) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.052079 (rank : 42) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.052072 (rank : 43) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.051409 (rank : 44) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.051219 (rank : 45) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.050983 (rank : 46) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.050593 (rank : 47) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.050365 (rank : 48) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.050102 (rank : 49) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.048925 (rank : 50) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.048693 (rank : 51) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.048630 (rank : 52) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.048580 (rank : 53) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.048520 (rank : 54) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.048465 (rank : 55) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.047874 (rank : 56) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
AMRP_MOUSE
|
||||||
| NC score | 0.046893 (rank : 57) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55302 | Gene names | Lrpap1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin receptor-associated protein precursor (Alpha-2- MRAP) (Low density lipoprotein receptor-related protein-associated protein 1) (RAP) (Heparin-binding protein 44) (HBP-44). | |||||
|
LC7L2_MOUSE
|
||||||
| NC score | 0.046877 (rank : 58) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.046816 (rank : 59) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
LC7L2_HUMAN
|
||||||
| NC score | 0.046724 (rank : 60) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.045237 (rank : 61) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.043936 (rank : 62) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
AATC_HUMAN
|
||||||
| NC score | 0.042608 (rank : 63) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17174 | Gene names | GOT1 | |||
|
Domain Architecture |

|
|||||
| Description | Aspartate aminotransferase, cytoplasmic (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 1). | |||||
|
RILP_HUMAN
|
||||||
| NC score | 0.042362 (rank : 64) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96NA2, Q71RE6, Q9BSL3, Q9BYS3 | Gene names | RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab-interacting lysosomal protein. | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.041781 (rank : 65) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.041622 (rank : 66) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.041449 (rank : 67) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
LAMA4_MOUSE
|
||||||
| NC score | 0.041191 (rank : 68) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
MAF_MOUSE
|
||||||
| NC score | 0.040532 (rank : 69) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
RAB3I_HUMAN
|
||||||
| NC score | 0.039889 (rank : 70) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.038874 (rank : 71) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.038708 (rank : 72) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MAF_HUMAN
|
||||||
| NC score | 0.038258 (rank : 73) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
AMRP_HUMAN
|
||||||
| NC score | 0.036966 (rank : 74) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30533 | Gene names | LRPAP1, A2MRAP | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin receptor-associated protein precursor (Alpha-2- MRAP) (Low density lipoprotein receptor-related protein-associated protein 1) (RAP). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.034026 (rank : 75) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.033204 (rank : 76) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
AATC_MOUSE
|
||||||
| NC score | 0.032461 (rank : 77) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05201 | Gene names | Got1 | |||
|
Domain Architecture |

|
|||||
| Description | Aspartate aminotransferase, cytoplasmic (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 1). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.031360 (rank : 78) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.025490 (rank : 79) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.024811 (rank : 80) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.023713 (rank : 81) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
CAB45_HUMAN
|
||||||
| NC score | 0.021154 (rank : 82) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRK5, Q8NBQ3, Q9NZP7 | Gene names | SDF4, CAB45 | |||
|
Domain Architecture |

|
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.019522 (rank : 83) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.019395 (rank : 84) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.018100 (rank : 85) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.017936 (rank : 86) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
SEPT8_HUMAN
|
||||||
| NC score | 0.012632 (rank : 87) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92599, Q8IX36, Q8IX37, Q9BVB3 | Gene names | SEPT8, KIAA0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
SEPT8_MOUSE
|
||||||
| NC score | 0.012078 (rank : 88) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CHH9, Q80YC7, Q9ESF7 | Gene names | Sept8, Kiaa0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.008999 (rank : 89) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
FER_HUMAN
|
||||||
| NC score | 0.006382 (rank : 90) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P16591 | Gene names | FER, TYK3 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FER (EC 2.7.10.2) (p94-FER) (c- FER). | |||||