Please be patient as the page loads
|
LMA1L_HUMAN
|
||||||
| SwissProt Accessions | Q9HAT1, Q6UWN2 | Gene names | LMAN1L, ERGL | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LMA1L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAT1, Q6UWN2 | Gene names | LMAN1L, ERGL | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMA1L_MOUSE
|
||||||
| θ value | 3.5201e-129 (rank : 2) | NC score | 0.987070 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCD3 | Gene names | Lman1l, Ergl | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMAN1_HUMAN
|
||||||
| θ value | 7.97034e-73 (rank : 3) | NC score | 0.920505 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
LMAN1_MOUSE
|
||||||
| θ value | 1.31681e-67 (rank : 4) | NC score | 0.917470 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
LMAN2_MOUSE
|
||||||
| θ value | 1.4233e-29 (rank : 5) | NC score | 0.827328 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBH5, Q9CXG7 | Gene names | Lman2 | |||
|
Domain Architecture |
|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (Lectin, mannose- binding 2). | |||||
|
LMAN2_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 6) | NC score | 0.826045 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12907, Q53HH1 | Gene names | LMAN2, C5orf8 | |||
|
Domain Architecture |
|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (GP36b glycoprotein) (Lectin, mannose-binding 2). | |||||
|
LMA2L_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 7) | NC score | 0.808299 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0V9, Q53GV3, Q53S67, Q63HN6, Q8NBQ6, Q9BQ14 | Gene names | LMAN2L, VIPL | |||
|
Domain Architecture |
|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMA2L_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 8) | NC score | 0.806125 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59481, Q3TZ91 | Gene names | Lman2l, Vipl | |||
|
Domain Architecture |
|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.022464 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
HIRA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.019218 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |
|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
TMG2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.033346 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R182, Q9EQI2 | Gene names | Prrg2, N4wbp1 | |||
|
Domain Architecture |
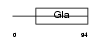
|
|||||
| Description | Transmembrane gamma-carboxyglutamic acid protein 2 precursor (Proline- rich Gla protein 2) (Proline-rich gamma-carboxyglutamic acid protein 2) (NEDD4 WW domain-binding protein 1). | |||||
|
M4A10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.029891 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N03, Q9D8H8, Q9EQY8 | Gene names | Ms4a10 | |||
|
Domain Architecture |
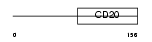
|
|||||
| Description | Membrane-spanning 4-domains subfamily A member 10. | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.000911 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
PDCD7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.018978 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
CRF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.024808 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06850 | Gene names | CRH | |||
|
Domain Architecture |

|
|||||
| Description | Corticoliberin precursor (Corticotropin-releasing factor) (CRF) (Corticotropin-releasing hormone). | |||||
|
NUCB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.009377 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.002338 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.013450 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.016863 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
GTR3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.005192 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32037 | Gene names | Slc2a3, Glut-3, Glut3 | |||
|
Domain Architecture |
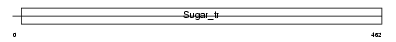
|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 3 (Glucose transporter type 3, brain). | |||||
|
LMA1L_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAT1, Q6UWN2 | Gene names | LMAN1L, ERGL | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMA1L_MOUSE
|
||||||
| NC score | 0.987070 (rank : 2) | θ value | 3.5201e-129 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCD3 | Gene names | Lman1l, Ergl | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMAN1_HUMAN
|
||||||
| NC score | 0.920505 (rank : 3) | θ value | 7.97034e-73 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
LMAN1_MOUSE
|
||||||
| NC score | 0.917470 (rank : 4) | θ value | 1.31681e-67 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
LMAN2_MOUSE
|
||||||
| NC score | 0.827328 (rank : 5) | θ value | 1.4233e-29 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBH5, Q9CXG7 | Gene names | Lman2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (Lectin, mannose- binding 2). | |||||
|
LMAN2_HUMAN
|
||||||
| NC score | 0.826045 (rank : 6) | θ value | 7.06379e-29 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12907, Q53HH1 | Gene names | LMAN2, C5orf8 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (GP36b glycoprotein) (Lectin, mannose-binding 2). | |||||
|
LMA2L_HUMAN
|
||||||
| NC score | 0.808299 (rank : 7) | θ value | 6.18819e-25 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0V9, Q53GV3, Q53S67, Q63HN6, Q8NBQ6, Q9BQ14 | Gene names | LMAN2L, VIPL | |||
|
Domain Architecture |

|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMA2L_MOUSE
|
||||||
| NC score | 0.806125 (rank : 8) | θ value | 6.84181e-24 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59481, Q3TZ91 | Gene names | Lman2l, Vipl | |||
|
Domain Architecture |

|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
TMG2_MOUSE
|
||||||
| NC score | 0.033346 (rank : 9) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R182, Q9EQI2 | Gene names | Prrg2, N4wbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane gamma-carboxyglutamic acid protein 2 precursor (Proline- rich Gla protein 2) (Proline-rich gamma-carboxyglutamic acid protein 2) (NEDD4 WW domain-binding protein 1). | |||||
|
M4A10_MOUSE
|
||||||
| NC score | 0.029891 (rank : 10) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N03, Q9D8H8, Q9EQY8 | Gene names | Ms4a10 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-spanning 4-domains subfamily A member 10. | |||||
|
CRF_HUMAN
|
||||||
| NC score | 0.024808 (rank : 11) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06850 | Gene names | CRH | |||
|
Domain Architecture |

|
|||||
| Description | Corticoliberin precursor (Corticotropin-releasing factor) (CRF) (Corticotropin-releasing hormone). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.022464 (rank : 12) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
HIRA_HUMAN
|
||||||
| NC score | 0.019218 (rank : 13) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |

|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
PDCD7_HUMAN
|
||||||
| NC score | 0.018978 (rank : 14) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.016863 (rank : 15) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.013450 (rank : 16) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
NUCB1_MOUSE
|
||||||
| NC score | 0.009377 (rank : 17) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
GTR3_MOUSE
|
||||||
| NC score | 0.005192 (rank : 18) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32037 | Gene names | Slc2a3, Glut-3, Glut3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 3 (Glucose transporter type 3, brain). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.002338 (rank : 19) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.000911 (rank : 20) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||