Please be patient as the page loads
|
LMAN1_MOUSE
|
||||||
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LMAN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988946 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
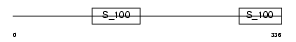
LMAN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
LMA1L_HUMAN
|
||||||
| θ value | 1.31681e-67 (rank : 3) | NC score | 0.917470 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HAT1, Q6UWN2 | Gene names | LMAN1L, ERGL | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMA1L_MOUSE
|
||||||
| θ value | 4.10295e-61 (rank : 4) | NC score | 0.921364 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VCD3 | Gene names | Lman1l, Ergl | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMAN2_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 5) | NC score | 0.853591 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBH5, Q9CXG7 | Gene names | Lman2 | |||
|
Domain Architecture |
|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (Lectin, mannose- binding 2). | |||||
|
LMAN2_HUMAN
|
||||||
| θ value | 5.21438e-40 (rank : 6) | NC score | 0.852722 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12907, Q53HH1 | Gene names | LMAN2, C5orf8 | |||
|
Domain Architecture |
|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (GP36b glycoprotein) (Lectin, mannose-binding 2). | |||||
|
LMA2L_HUMAN
|
||||||
| θ value | 6.37413e-38 (rank : 7) | NC score | 0.841780 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0V9, Q53GV3, Q53S67, Q63HN6, Q8NBQ6, Q9BQ14 | Gene names | LMAN2L, VIPL | |||
|
Domain Architecture |
|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMA2L_MOUSE
|
||||||
| θ value | 1.42001e-37 (rank : 8) | NC score | 0.840766 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59481, Q3TZ91 | Gene names | Lman2l, Vipl | |||
|
Domain Architecture |
|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
TUFT1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 9) | NC score | 0.079285 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.082877 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.068530 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.064301 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.069303 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.061343 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
K2C1B_MOUSE
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.025917 (rank : 92) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6IFZ6 | Gene names | Krt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b (Type II keratin Kb39) (Embryonic type II keratin-1). | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.068511 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.068218 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
LC7L2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.061325 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
LC7L2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.061608 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
AATC_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.060019 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17174 | Gene names | GOT1 | |||
|
Domain Architecture |
|
|||||
| Description | Aspartate aminotransferase, cytoplasmic (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 1). | |||||
|
LAMA4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.042241 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
AATC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.049956 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05201 | Gene names | Got1 | |||
|
Domain Architecture |
|
|||||
| Description | Aspartate aminotransferase, cytoplasmic (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 1). | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.056568 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.066078 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.072103 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.064692 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.057546 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.057307 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.059129 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.056435 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERCC5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.050901 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.030872 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.049282 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.048132 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
RM01_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.033245 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYD6, Q4W5B8, Q6IAG4, Q96BW3, Q9H793, Q9NRL5 | Gene names | MRPL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L1, mitochondrial precursor (L1mt) (MRP-L1). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.053316 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.052136 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.052885 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.053421 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.052525 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.026817 (rank : 91) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.061104 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.060230 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.046844 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
KIF23_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.047932 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.050490 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GCC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.054391 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.055276 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
MAF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.036166 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.043008 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.043134 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.036719 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.017646 (rank : 95) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
STX10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.029468 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60499, Q96AE8 | Gene names | STX10, SYN10 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-10 (Syn10). | |||||
|
CING_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.055157 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MAF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.033108 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.039571 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.056005 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
AMRP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.044895 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55302 | Gene names | Lrpap1 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2-macroglobulin receptor-associated protein precursor (Alpha-2- MRAP) (Low density lipoprotein receptor-related protein-associated protein 1) (RAP) (Heparin-binding protein 44) (HBP-44). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.045587 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
KINH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.057843 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.052016 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.051363 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.038719 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
ES8L3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.007639 (rank : 100) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.055388 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.058489 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.028838 (rank : 90) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.042469 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.054626 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
RFA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.019470 (rank : 93) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27694, Q59ES9 | Gene names | RPA1, REPA1, RPA70 | |||
|
Domain Architecture |
|
|||||
| Description | Replication protein A 70 kDa DNA-binding subunit (RP-A) (RF-A) (Replication factor-A protein 1) (Single-stranded DNA-binding protein) (p70). | |||||
|
TESC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.011921 (rank : 98) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKL5 | Gene names | Tesc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tescalcin (TSC). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.042485 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
AMRP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.036281 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P30533 | Gene names | LRPAP1, A2MRAP | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2-macroglobulin receptor-associated protein precursor (Alpha-2- MRAP) (Low density lipoprotein receptor-related protein-associated protein 1) (RAP). | |||||
|
CAB45_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.017324 (rank : 96) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRK5, Q8NBQ3, Q9NZP7 | Gene names | SDF4, CAB45 | |||
|
Domain Architecture |
|
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.051122 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CING_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.051631 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.045815 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.050028 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.038713 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DP13A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.010854 (rank : 99) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.054971 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.048943 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
LIN9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.014349 (rank : 97) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C735, Q5TKA0, Q8C9D8 | Gene names | Lin9, Bara, Tgs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-9 homolog (mLin-9) (Type I interferon receptor beta chain- associated protein) (TUDOR gene similar 1 protein). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.039148 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.041655 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
RFA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.018161 (rank : 94) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEE4, Q3TEJ8 | Gene names | Rpa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication protein A 70 kDa DNA-binding subunit (RP-A) (RF-A) (Replication factor-A protein 1) (p70). | |||||
|
C102B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050230 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
CCD27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.056231 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.052557 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
EP15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.053587 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.057031 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.052259 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.054801 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.053965 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.050538 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.050408 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050374 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.051124 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TUFT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.053585 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
LMAN1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
LMAN1_HUMAN
|
||||||
| NC score | 0.988946 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
LMA1L_MOUSE
|
||||||
| NC score | 0.921364 (rank : 3) | θ value | 4.10295e-61 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VCD3 | Gene names | Lman1l, Ergl | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMA1L_HUMAN
|
||||||
| NC score | 0.917470 (rank : 4) | θ value | 1.31681e-67 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HAT1, Q6UWN2 | Gene names | LMAN1L, ERGL | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMAN2_MOUSE
|
||||||
| NC score | 0.853591 (rank : 5) | θ value | 1.05066e-40 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBH5, Q9CXG7 | Gene names | Lman2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (Lectin, mannose- binding 2). | |||||
|
LMAN2_HUMAN
|
||||||
| NC score | 0.852722 (rank : 6) | θ value | 5.21438e-40 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12907, Q53HH1 | Gene names | LMAN2, C5orf8 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (GP36b glycoprotein) (Lectin, mannose-binding 2). | |||||
|
LMA2L_HUMAN
|
||||||
| NC score | 0.841780 (rank : 7) | θ value | 6.37413e-38 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0V9, Q53GV3, Q53S67, Q63HN6, Q8NBQ6, Q9BQ14 | Gene names | LMAN2L, VIPL | |||
|
Domain Architecture |

|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMA2L_MOUSE
|
||||||
| NC score | 0.840766 (rank : 8) | θ value | 1.42001e-37 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59481, Q3TZ91 | Gene names | Lman2l, Vipl | |||
|
Domain Architecture |

|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.082877 (rank : 9) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
TUFT1_MOUSE
|
||||||
| NC score | 0.079285 (rank : 10) | θ value | 0.0148317 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.072103 (rank : 11) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.069303 (rank : 12) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.068530 (rank : 13) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.068511 (rank : 14) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.068218 (rank : 15) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.066078 (rank : 16) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.064692 (rank : 17) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.064301 (rank : 18) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
LC7L2_MOUSE
|
||||||
| NC score | 0.061608 (rank : 19) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.061343 (rank : 20) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
LC7L2_HUMAN
|
||||||
| NC score | 0.061325 (rank : 21) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.061104 (rank : 22) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.060230 (rank : 23) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
AATC_HUMAN
|
||||||
| NC score | 0.060019 (rank : 24) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17174 | Gene names | GOT1 | |||
|
Domain Architecture |

|
|||||
| Description | Aspartate aminotransferase, cytoplasmic (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 1). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.059129 (rank : 25) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.058489 (rank : 26) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.057843 (rank : 27) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.057546 (rank : 28) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.057307 (rank : 29) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.057031 (rank : 30) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.056568 (rank : 31) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.056435 (rank : 32) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
CCD27_HUMAN
|
||||||
| NC score | 0.056231 (rank : 33) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.056005 (rank : 34) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.055388 (rank : 35) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.055276 (rank : 36) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.055157 (rank : 37) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.054971 (rank : 38) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.054801 (rank : 39) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.054626 (rank : 40) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
GCC1_HUMAN
|
||||||
| NC score | 0.054391 (rank : 41) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.053965 (rank : 42) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.053587 (rank : 43) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
TUFT1_HUMAN
|
||||||
| NC score | 0.053585 (rank : 44) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.053421 (rank : 45) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.053316 (rank : 46) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.052885 (rank : 47) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.052557 (rank : 48) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.052525 (rank : 49) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.052259 (rank : 50) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.052136 (rank : 51) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.052016 (rank : 52) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.051631 (rank : 53) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.051363 (rank : 54) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.051124 (rank : 55) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.051122 (rank : 56) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.050901 (rank : 57) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.050538 (rank : 58) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.050490 (rank : 59) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.050408 (rank : 60) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.050374 (rank : 61) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
C102B_HUMAN
|
||||||
| NC score | 0.050230 (rank : 62) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.050028 (rank : 63) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
AATC_MOUSE
|
||||||
| NC score | 0.049956 (rank : 64) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05201 | Gene names | Got1 | |||
|
Domain Architecture |

|
|||||
| Description | Aspartate aminotransferase, cytoplasmic (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 1). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.049282 (rank : 65) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.048943 (rank : 66) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.048132 (rank : 67) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
KIF23_HUMAN
|
||||||
| NC score | 0.047932 (rank : 68) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.046844 (rank : 69) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.045815 (rank : 70) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.045587 (rank : 71) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
AMRP_MOUSE
|
||||||
| NC score | 0.044895 (rank : 72) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55302 | Gene names | Lrpap1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin receptor-associated protein precursor (Alpha-2- MRAP) (Low density lipoprotein receptor-related protein-associated protein 1) (RAP) (Heparin-binding protein 44) (HBP-44). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.043134 (rank : 73) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.043008 (rank : 74) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.042485 (rank : 75) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.042469 (rank : 76) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
LAMA4_MOUSE
|
||||||
| NC score | 0.042241 (rank : 77) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.041655 (rank : 78) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.039571 (rank : 79) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.039148 (rank : 80) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.038719 (rank : 81) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.038713 (rank : 82) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.036719 (rank : 83) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
AMRP_HUMAN
|
||||||
| NC score | 0.036281 (rank : 84) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P30533 | Gene names | LRPAP1, A2MRAP | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin receptor-associated protein precursor (Alpha-2- MRAP) (Low density lipoprotein receptor-related protein-associated protein 1) (RAP). | |||||
|
MAF_MOUSE
|
||||||
| NC score | 0.036166 (rank : 85) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
RM01_HUMAN
|
||||||
| NC score | 0.033245 (rank : 86) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYD6, Q4W5B8, Q6IAG4, Q96BW3, Q9H793, Q9NRL5 | Gene names | MRPL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L1, mitochondrial precursor (L1mt) (MRP-L1). | |||||
|
MAF_HUMAN
|
||||||
| NC score | 0.033108 (rank : 87) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.030872 (rank : 88) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
STX10_HUMAN
|
||||||
| NC score | 0.029468 (rank : 89) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60499, Q96AE8 | Gene names | STX10, SYN10 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-10 (Syn10). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.028838 (rank : 90) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.026817 (rank : 91) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
K2C1B_MOUSE
|
||||||
| NC score | 0.025917 (rank : 92) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6IFZ6 | Gene names | Krt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b (Type II keratin Kb39) (Embryonic type II keratin-1). | |||||
|
RFA1_HUMAN
|
||||||
| NC score | 0.019470 (rank : 93) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27694, Q59ES9 | Gene names | RPA1, REPA1, RPA70 | |||
|
Domain Architecture |

|
|||||
| Description | Replication protein A 70 kDa DNA-binding subunit (RP-A) (RF-A) (Replication factor-A protein 1) (Single-stranded DNA-binding protein) (p70). | |||||
|
RFA1_MOUSE
|
||||||
| NC score | 0.018161 (rank : 94) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEE4, Q3TEJ8 | Gene names | Rpa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication protein A 70 kDa DNA-binding subunit (RP-A) (RF-A) (Replication factor-A protein 1) (p70). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.017646 (rank : 95) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CAB45_HUMAN
|
||||||
| NC score | 0.017324 (rank : 96) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRK5, Q8NBQ3, Q9NZP7 | Gene names | SDF4, CAB45 | |||
|
Domain Architecture |

|
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
LIN9_MOUSE
|
||||||
| NC score | 0.014349 (rank : 97) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C735, Q5TKA0, Q8C9D8 | Gene names | Lin9, Bara, Tgs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-9 homolog (mLin-9) (Type I interferon receptor beta chain- associated protein) (TUDOR gene similar 1 protein). | |||||
|
TESC_MOUSE
|
||||||
| NC score | 0.011921 (rank : 98) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKL5 | Gene names | Tesc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tescalcin (TSC). | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.010854 (rank : 99) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
ES8L3_HUMAN
|
||||||
| NC score | 0.007639 (rank : 100) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||