Please be patient as the page loads
|
CABYR_HUMAN
|
||||||
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CABYR_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
CABYR_MOUSE
|
||||||
| θ value | 9.89713e-132 (rank : 2) | NC score | 0.916599 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
SP17_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 3) | NC score | 0.394410 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62252 | Gene names | Spa17, Sp17 | |||
|
Domain Architecture |
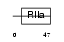
|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 4) | NC score | 0.112175 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SP17_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 5) | NC score | 0.355483 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15506, Q9BXF7 | Gene names | SPA17, SP17 | |||
|
Domain Architecture |
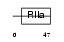
|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17) (Sperm protein 17) (Sp17-1). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 6) | NC score | 0.103073 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.072103 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.091525 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.042721 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
AR13B_MOUSE
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.033268 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q640N2 | Gene names | Arl13b, Arl2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.070918 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.040721 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.034619 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.037676 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.030216 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.030592 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
INVO_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.038247 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
TAF9B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.053216 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBM6, Q9Y2S3 | Gene names | TAF9B, TAF9L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 9B (Transcription initiation factor TFIID subunit 9-like protein) (Transcription- associated factor TAFII31L) (Neuronal cell death-related protein 7) (DN-7). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.058579 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.042547 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TM1L1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.063359 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.003506 (rank : 91) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.028041 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
ERCC5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.052670 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.021208 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.039157 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.057826 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.055778 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.029730 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZEP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.029010 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
DNAI1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.035335 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UI46, Q9UEZ8 | Gene names | DNAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein intermediate chain 1, axonemal (Axonemal dynein intermediate chain 1). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.031603 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.050325 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.046872 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.050803 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.049629 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.041148 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
RANB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.043820 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H6Z4, O60405, O75759, O75760, Q9BT47, Q9UG74 | Gene names | RANBP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.026000 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CRX_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.010412 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54751 | Gene names | Crx | |||
|
Domain Architecture |
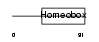
|
|||||
| Description | Cone-rod homeobox protein. | |||||
|
GCC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.023657 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.022463 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
PAK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.003803 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13177, Q13154 | Gene names | PAK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 2 (EC 2.7.11.1) (p21-activated kinase 2) (PAK-2) (PAK65) (Gamma-PAK) (S6/H4 kinase). | |||||
|
DCDC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.042322 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |
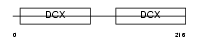
|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
KAP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.052060 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.025517 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.040955 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TF3C1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.028670 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K284, Q8CAL9 | Gene names | Gtf3c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.026447 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.025788 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.003645 (rank : 90) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14757 | Gene names | CHEK1, CHK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Chk1 (EC 2.7.11.1). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.011711 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
KAP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.036639 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
LIPB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.014865 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
NUAK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.003053 (rank : 92) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||
|
SCG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.032655 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.019672 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.036039 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.035312 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.022854 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.025528 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.032495 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.011573 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
REPS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.021246 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.046451 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.026303 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TXD13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.020353 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.014741 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
ZWINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.022427 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.012577 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.034438 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
TAF9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.036350 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VI33, Q32P09, Q80XS3, Q9CV61 | Gene names | Taf9, Taf2g | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 9 (Transcription initiation factor TFIID 31 kDa subunit) (TAFII-31) (TAFII-32) (TAFII32). | |||||
|
VMDL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.013282 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N1M1, Q8N356, Q8NFT9, Q9BR80 | Gene names | VMD2L3 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-4 (Vitelliform macular dystrophy 2-like protein 3). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.021391 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.012414 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.010909 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IP6K1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.010988 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92551, Q7L3I7, Q96E38 | Gene names | IHPK1, IP6K1, KIAA0263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol hexaphosphate kinase 1 (EC 2.7.4.21) (InsP6 kinase 1) (Inositol hexakisphosphate kinase 1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.015833 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.063644 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.033204 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.025311 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MPRI_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.012672 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.042781 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RABE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.014508 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |
|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.016194 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
UCHL4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.011936 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58321 | Gene names | Uchl4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L4 (EC 3.4.19.12) (UCH- L4) (Ubiquitin thioesterase L4). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.025012 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053983 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.059560 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
MINT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.052975 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RP1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.057633 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.056837 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CABYR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
CABYR_MOUSE
|
||||||
| NC score | 0.916599 (rank : 2) | θ value | 9.89713e-132 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
SP17_MOUSE
|
||||||
| NC score | 0.394410 (rank : 3) | θ value | 1.29631e-06 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62252 | Gene names | Spa17, Sp17 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17). | |||||
|
SP17_HUMAN
|
||||||
| NC score | 0.355483 (rank : 4) | θ value | 9.29e-05 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15506, Q9BXF7 | Gene names | SPA17, SP17 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17) (Sperm protein 17) (Sp17-1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.112175 (rank : 5) | θ value | 3.19293e-05 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.103073 (rank : 6) | θ value | 0.00228821 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.091525 (rank : 7) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.072103 (rank : 8) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.070918 (rank : 9) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.063644 (rank : 10) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TM1L1_HUMAN
|
||||||
| NC score | 0.063359 (rank : 11) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.059560 (rank : 12) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.058579 (rank : 13) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.057826 (rank : 14) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
RP1L1_MOUSE
|
||||||
| NC score | 0.057633 (rank : 15) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.056837 (rank : 16) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.055778 (rank : 17) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.053983 (rank : 18) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TAF9B_HUMAN
|
||||||
| NC score | 0.053216 (rank : 19) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBM6, Q9Y2S3 | Gene names | TAF9B, TAF9L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 9B (Transcription initiation factor TFIID subunit 9-like protein) (Transcription- associated factor TAFII31L) (Neuronal cell death-related protein 7) (DN-7). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.052975 (rank : 20) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.052670 (rank : 21) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.052060 (rank : 22) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.050803 (rank : 23) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.050325 (rank : 24) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.049629 (rank : 25) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.046872 (rank : 26) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.046451 (rank : 27) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
RANB3_HUMAN
|
||||||
| NC score | 0.043820 (rank : 28) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H6Z4, O60405, O75759, O75760, Q9BT47, Q9UG74 | Gene names | RANBP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.042781 (rank : 29) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.042721 (rank : 30) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.042547 (rank : 31) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
DCDC2_HUMAN
|
||||||
| NC score | 0.042322 (rank : 32) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |

|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.041148 (rank : 33) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.040955 (rank : 34) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.040721 (rank : 35) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.039157 (rank : 36) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.038247 (rank : 37) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.037676 (rank : 38) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.036639 (rank : 39) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
TAF9_MOUSE
|
||||||
| NC score | 0.036350 (rank : 40) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VI33, Q32P09, Q80XS3, Q9CV61 | Gene names | Taf9, Taf2g | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 9 (Transcription initiation factor TFIID 31 kDa subunit) (TAFII-31) (TAFII-32) (TAFII32). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.036039 (rank : 41) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
DNAI1_HUMAN
|
||||||
| NC score | 0.035335 (rank : 42) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UI46, Q9UEZ8 | Gene names | DNAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein intermediate chain 1, axonemal (Axonemal dynein intermediate chain 1). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.035312 (rank : 43) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.034619 (rank : 44) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.034438 (rank : 45) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
AR13B_MOUSE
|
||||||
| NC score | 0.033268 (rank : 46) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q640N2 | Gene names | Arl13b, Arl2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.033204 (rank : 47) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
SCG1_HUMAN
|
||||||
| NC score | 0.032655 (rank : 48) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.032495 (rank : 49) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.031603 (rank : 50) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.030592 (rank : 51) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.030216 (rank : 52) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.029730 (rank : 53) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZEP2_HUMAN
|
||||||
| NC score | 0.029010 (rank : 54) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
TF3C1_MOUSE
|
||||||
| NC score | 0.028670 (rank : 55) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K284, Q8CAL9 | Gene names | Gtf3c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.028041 (rank : 56) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.026447 (rank : 57) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.026303 (rank : 58) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.026000 (rank : 59) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.025788 (rank : 60) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.025528 (rank : 61) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.025517 (rank : 62) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.025311 (rank : 63) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.025012 (rank : 64) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
GCC1_HUMAN
|
||||||
| NC score | 0.023657 (rank : 65) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.022854 (rank : 66) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.022463 (rank : 67) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
ZWINT_MOUSE
|
||||||
| NC score | 0.022427 (rank : 68) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.021391 (rank : 69) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
REPS2_HUMAN
|
||||||
| NC score | 0.021246 (rank : 70) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.021208 (rank : 71) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TXD13_HUMAN
|
||||||
| NC score | 0.020353 (rank : 72) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.019672 (rank : 73) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.016194 (rank : 74) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.015833 (rank : 75) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
LIPB1_HUMAN
|
||||||
| NC score | 0.014865 (rank : 76) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.014741 (rank : 77) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
RABE2_HUMAN
|
||||||
| NC score | 0.014508 (rank : 78) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
VMDL3_HUMAN
|
||||||
| NC score | 0.013282 (rank : 79) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N1M1, Q8N356, Q8NFT9, Q9BR80 | Gene names | VMD2L3 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-4 (Vitelliform macular dystrophy 2-like protein 3). | |||||
|
MPRI_MOUSE
|
||||||
| NC score | 0.012672 (rank : 80) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.012577 (rank : 81) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.012414 (rank : 82) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
UCHL4_MOUSE
|
||||||
| NC score | 0.011936 (rank : 83) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58321 | Gene names | Uchl4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L4 (EC 3.4.19.12) (UCH- L4) (Ubiquitin thioesterase L4). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.011711 (rank : 84) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.011573 (rank : 85) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
IP6K1_HUMAN
|
||||||
| NC score | 0.010988 (rank : 86) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92551, Q7L3I7, Q96E38 | Gene names | IHPK1, IP6K1, KIAA0263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol hexaphosphate kinase 1 (EC 2.7.4.21) (InsP6 kinase 1) (Inositol hexakisphosphate kinase 1). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.010909 (rank : 87) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
CRX_MOUSE
|
||||||
| NC score | 0.010412 (rank : 88) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54751 | Gene names | Crx | |||
|
Domain Architecture |

|
|||||
| Description | Cone-rod homeobox protein. | |||||
|
PAK2_HUMAN
|
||||||
| NC score | 0.003803 (rank : 89) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13177, Q13154 | Gene names | PAK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 2 (EC 2.7.11.1) (p21-activated kinase 2) (PAK-2) (PAK65) (Gamma-PAK) (S6/H4 kinase). | |||||
|
CHK1_HUMAN
|
||||||
| NC score | 0.003645 (rank : 90) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14757 | Gene names | CHEK1, CHK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Chk1 (EC 2.7.11.1). | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | 0.003506 (rank : 91) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
NUAK1_HUMAN
|
||||||
| NC score | 0.003053 (rank : 92) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||