Please be patient as the page loads
|
SP17_HUMAN
|
||||||
| SwissProt Accessions | Q15506, Q9BXF7 | Gene names | SPA17, SP17 | |||
|
Domain Architecture |
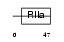
|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17) (Sperm protein 17) (Sp17-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SP17_HUMAN
|
||||||
| θ value | 1.7756e-72 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15506, Q9BXF7 | Gene names | SPA17, SP17 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17) (Sperm protein 17) (Sp17-1). | |||||
|
SP17_MOUSE
|
||||||
| θ value | 1.23823e-49 (rank : 2) | NC score | 0.972040 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62252 | Gene names | Spa17, Sp17 | |||
|
Domain Architecture |
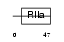
|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17). | |||||
|
CABYR_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 3) | NC score | 0.355483 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
KAP2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 4) | NC score | 0.226644 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
CABYR_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 5) | NC score | 0.325482 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
KAP3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.189033 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.188571 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.138655 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.062499 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.061563 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
KAP2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.179933 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
DPOLZ_MOUSE
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.048651 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
MYO3A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.011981 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||
|
OFD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.027874 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.089216 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
GALT5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.013273 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.034941 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.034268 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
FA13A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.024706 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
LRC27_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.019936 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
NEUG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.157906 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92686 | Gene names | NRGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogranin (Ng) (RC3). | |||||
|
NEUG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.151734 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60761, Q3TYH4 | Gene names | Nrgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogranin (Ng) (RC3). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.020910 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
HNRPQ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.027025 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |
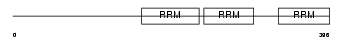
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
HNRPQ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.027139 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |
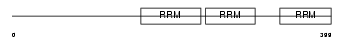
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.011690 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.021896 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
ADA30_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.007562 (rank : 37) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
IQGA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.015629 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.015101 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
LDB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.024616 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U70, O96010, Q9UGM4 | Gene names | LDB1, CLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 1 (Carboxyl-terminal LIM domain-binding protein 2) (CLIM-2) (LIM domain-binding factor CLIM2). | |||||
|
LDB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.024612 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70662 | Gene names | Ldb1, Nli | |||
|
Domain Architecture |
|
|||||
| Description | LIM domain-binding protein 1 (Nuclear LIM interactor). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.002824 (rank : 38) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.049446 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
KAP0_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.093655 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.094153 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.094123 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.094281 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
SP17_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.7756e-72 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15506, Q9BXF7 | Gene names | SPA17, SP17 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17) (Sperm protein 17) (Sp17-1). | |||||
|
SP17_MOUSE
|
||||||
| NC score | 0.972040 (rank : 2) | θ value | 1.23823e-49 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62252 | Gene names | Spa17, Sp17 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17). | |||||
|
CABYR_HUMAN
|
||||||
| NC score | 0.355483 (rank : 3) | θ value | 9.29e-05 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
CABYR_MOUSE
|
||||||
| NC score | 0.325482 (rank : 4) | θ value | 0.000786445 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.226644 (rank : 5) | θ value | 9.29e-05 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.189033 (rank : 6) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.188571 (rank : 7) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.179933 (rank : 8) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
NEUG_HUMAN
|
||||||
| NC score | 0.157906 (rank : 9) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92686 | Gene names | NRGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogranin (Ng) (RC3). | |||||
|
NEUG_MOUSE
|
||||||
| NC score | 0.151734 (rank : 10) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60761, Q3TYH4 | Gene names | Nrgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogranin (Ng) (RC3). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.138655 (rank : 11) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.094281 (rank : 12) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.094153 (rank : 13) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.094123 (rank : 14) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.093655 (rank : 15) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.089216 (rank : 16) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.062499 (rank : 17) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.061563 (rank : 18) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.049446 (rank : 19) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
DPOLZ_MOUSE
|
||||||
| NC score | 0.048651 (rank : 20) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.034941 (rank : 21) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.034268 (rank : 22) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.027874 (rank : 23) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
HNRPQ_MOUSE
|
||||||
| NC score | 0.027139 (rank : 24) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
HNRPQ_HUMAN
|
||||||
| NC score | 0.027025 (rank : 25) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.024706 (rank : 26) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
LDB1_HUMAN
|
||||||
| NC score | 0.024616 (rank : 27) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U70, O96010, Q9UGM4 | Gene names | LDB1, CLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 1 (Carboxyl-terminal LIM domain-binding protein 2) (CLIM-2) (LIM domain-binding factor CLIM2). | |||||
|
LDB1_MOUSE
|
||||||
| NC score | 0.024612 (rank : 28) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70662 | Gene names | Ldb1, Nli | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-binding protein 1 (Nuclear LIM interactor). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.021896 (rank : 29) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.020910 (rank : 30) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
LRC27_HUMAN
|
||||||
| NC score | 0.019936 (rank : 31) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
IQGA1_HUMAN
|
||||||
| NC score | 0.015629 (rank : 32) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.015101 (rank : 33) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
GALT5_HUMAN
|
||||||
| NC score | 0.013273 (rank : 34) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
MYO3A_HUMAN
|
||||||
| NC score | 0.011981 (rank : 35) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.011690 (rank : 36) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
ADA30_HUMAN
|
||||||
| NC score | 0.007562 (rank : 37) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.002824 (rank : 38) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||