Please be patient as the page loads
|
EDEM3_HUMAN
|
||||||
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EDEM3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
EDEM1_HUMAN
|
||||||
| θ value | 1.43912e-106 (rank : 2) | NC score | 0.929976 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM1_MOUSE
|
||||||
| θ value | 1.43912e-106 (rank : 3) | NC score | 0.928802 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q925U4 | Gene names | Edem1, Edem | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM2_HUMAN
|
||||||
| θ value | 2.47191e-82 (rank : 4) | NC score | 0.927857 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BV94, Q6IA89, Q6UWZ4, Q9H4U0, Q9H886, Q9NTL9, Q9NVE6 | Gene names | EDEM2, C20orf31, C20orf49 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 2 precursor. | |||||
|
MA1B1_HUMAN
|
||||||
| θ value | 2.25659e-51 (rank : 5) | NC score | 0.850558 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
MA1A1_HUMAN
|
||||||
| θ value | 3.98324e-48 (rank : 6) | NC score | 0.832578 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33908, Q9NU44, Q9UJI3 | Gene names | MAN1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1A1_MOUSE
|
||||||
| θ value | 7.51208e-47 (rank : 7) | NC score | 0.831835 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P45700 | Gene names | Man1a1, Man1a | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1A2_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 8) | NC score | 0.824776 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60476, Q9H510 | Gene names | MAN1A2, MAN1B | |||
|
Domain Architecture |
|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1A2_MOUSE
|
||||||
| θ value | 9.20422e-37 (rank : 9) | NC score | 0.821882 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39098, Q5SUY6, Q5SUY7, Q60599, Q9CUY9 | Gene names | Man1a2, Man1b | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1C1_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 10) | NC score | 0.829305 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR34, Q9Y545 | Gene names | MAN1C1, MAN1A3, MAN1C | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IC) (Alpha-1,2-mannosidase IC) (Mannosidase alpha class 1C member 1) (HMIC). | |||||
|
PAP21_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 11) | NC score | 0.200445 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BSG0 | Gene names | PAP21, C2orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protease-associated domain-containing protein of 21 kDa precursor (hPAP21). | |||||
|
PAP21_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 12) | NC score | 0.200492 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D9N8 | Gene names | Pap21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protease-associated domain-containing protein of 21 kDa precursor. | |||||
|
RNF13_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.113956 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54965, O54966 | Gene names | Rnf13, Rzf | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 13. | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.042511 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
RN167_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 15) | NC score | 0.108891 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H6Y7, Q6XYE0, Q8NDC1, Q9Y3V1 | Gene names | RNF167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 167 precursor. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 16) | NC score | 0.071940 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RNF13_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 17) | NC score | 0.108468 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43567 | Gene names | RNF13, RZF | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 13. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 18) | NC score | 0.084328 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
RN167_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 19) | NC score | 0.106663 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XF4, Q3U4S5 | Gene names | Rnf167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 167 precursor. | |||||
|
PSL1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.078480 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TD49, Q3TLQ8, Q3UG60, Q80VZ6 | Gene names | Sppl2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b). | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.033948 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.055312 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.057635 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
POT15_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.033451 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.012761 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PSL2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.071494 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TCT8, Q8TAW1, Q96SZ8 | Gene names | SPPL2A, PSL2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2A (EC 3.4.23.-) (Protein SPP-like 2A) (Protein SPPL2a) (Intramembrane protease 3) (IMP3) (Presenilin-like protein 2). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.039164 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.054323 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
RP1L1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.050785 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
TFR1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.032683 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62351, Q61560 | Gene names | Tfrc, Trfr | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen). | |||||
|
CB010_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.049939 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z570, Q6ZN26 | Gene names | C2orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf10. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.047679 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.054028 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.022605 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.065189 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.048209 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.057333 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RN133_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.086332 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WVZ7, Q8N7G7 | Gene names | RNF133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 133. | |||||
|
ZHX2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.021731 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6X8 | Gene names | ZHX2, AFR1, KIAA0854, RAF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.046441 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.011589 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.032008 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
GOLI_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.073448 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86XS8, Q641P9, Q6P177, Q7L2U2, Q9P0J9 | Gene names | RNF130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130). | |||||
|
GOLI_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.073419 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VEM1, Q80VL7, Q9QZQ6 | Gene names | Rnf130, G1rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130) (G1-related zinc finger protein). | |||||
|
CF051_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.050736 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969F1 | Gene names | C6orf51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf51. | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.016724 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.018957 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.044571 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.018352 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
RAE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.019172 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24386, O43732 | Gene names | CHM, REP1, TCD | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein) (TCD protein). | |||||
|
GCDH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.014879 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92947, O14719 | Gene names | GCDH | |||
|
Domain Architecture |
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.008304 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.040577 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
RPF53_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.027495 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K202, Q62034, Q8BQG9 | Gene names | Praf1, Paf53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase I-associated factor 53 kDa subunit (EC 2.7.7.6) (RNA polymerase I-associated factor 1). | |||||
|
SAS10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.036138 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.029807 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.019146 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.017037 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
ERCC5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.024600 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.008397 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.038333 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
FA29A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.026149 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
GP158_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.015715 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
POT14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.029951 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.034040 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.012796 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
SGOL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.018590 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TSY8, Q811I4, Q8C9C4, Q8CGJ4, Q9CS55 | Gene names | Sgol2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2. | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.034478 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.023238 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.003755 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.060148 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
EMR3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.003895 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY15 | Gene names | EMR3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 3 precursor (EGF-like module-containing mucin-like receptor EMR3). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.009668 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
MYPC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.004580 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XKE0, Q8C109, Q8K2V0 | Gene names | Mybpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.019737 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
SMBT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.010652 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHJ3, Q96C73, Q9Y4Q9 | Gene names | SFMBT1, RU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1 (Renal ubiquitous protein 1). | |||||
|
TFR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.023017 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02786, Q59G55, Q9UCN0, Q9UCU5, Q9UDF9, Q9UK21 | Gene names | TFRC | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen) (T9) (p90). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.032260 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.009306 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.009460 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
LRFN5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.001167 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA0, Q8BJH4, Q8BZL0 | Gene names | Lrfn5 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.029312 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.027614 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.015675 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.014309 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.008503 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
PPHLN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.013525 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2H1, Q8BUZ6 | Gene names | Pphln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1. | |||||
|
PSL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.056800 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJF9, Q8R354 | Gene names | Sppl2a, Psl2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2A (EC 3.4.23.-) (SPP-like 2A protein) (Protein SPPL2a) (Intramembrane protease 3) (IMP3) (Presenilin-like protein 2). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.010023 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.041044 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.011328 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.021824 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
EDEM3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
EDEM1_HUMAN
|
||||||
| NC score | 0.929976 (rank : 2) | θ value | 1.43912e-106 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM1_MOUSE
|
||||||
| NC score | 0.928802 (rank : 3) | θ value | 1.43912e-106 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q925U4 | Gene names | Edem1, Edem | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM2_HUMAN
|
||||||
| NC score | 0.927857 (rank : 4) | θ value | 2.47191e-82 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BV94, Q6IA89, Q6UWZ4, Q9H4U0, Q9H886, Q9NTL9, Q9NVE6 | Gene names | EDEM2, C20orf31, C20orf49 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 2 precursor. | |||||
|
MA1B1_HUMAN
|
||||||
| NC score | 0.850558 (rank : 5) | θ value | 2.25659e-51 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
MA1A1_HUMAN
|
||||||
| NC score | 0.832578 (rank : 6) | θ value | 3.98324e-48 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33908, Q9NU44, Q9UJI3 | Gene names | MAN1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1A1_MOUSE
|
||||||
| NC score | 0.831835 (rank : 7) | θ value | 7.51208e-47 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P45700 | Gene names | Man1a1, Man1a | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1C1_HUMAN
|
||||||
| NC score | 0.829305 (rank : 8) | θ value | 3.86705e-35 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR34, Q9Y545 | Gene names | MAN1C1, MAN1A3, MAN1C | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IC) (Alpha-1,2-mannosidase IC) (Mannosidase alpha class 1C member 1) (HMIC). | |||||
|
MA1A2_HUMAN
|
||||||
| NC score | 0.824776 (rank : 9) | θ value | 2.86122e-38 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60476, Q9H510 | Gene names | MAN1A2, MAN1B | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1A2_MOUSE
|
||||||
| NC score | 0.821882 (rank : 10) | θ value | 9.20422e-37 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39098, Q5SUY6, Q5SUY7, Q60599, Q9CUY9 | Gene names | Man1a2, Man1b | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
PAP21_MOUSE
|
||||||
| NC score | 0.200492 (rank : 11) | θ value | 2.88788e-06 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D9N8 | Gene names | Pap21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protease-associated domain-containing protein of 21 kDa precursor. | |||||
|
PAP21_HUMAN
|
||||||
| NC score | 0.200445 (rank : 12) | θ value | 2.88788e-06 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BSG0 | Gene names | PAP21, C2orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protease-associated domain-containing protein of 21 kDa precursor (hPAP21). | |||||
|
RNF13_MOUSE
|
||||||
| NC score | 0.113956 (rank : 13) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54965, O54966 | Gene names | Rnf13, Rzf | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 13. | |||||
|
RN167_HUMAN
|
||||||
| NC score | 0.108891 (rank : 14) | θ value | 0.00228821 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H6Y7, Q6XYE0, Q8NDC1, Q9Y3V1 | Gene names | RNF167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 167 precursor. | |||||
|
RNF13_HUMAN
|
||||||
| NC score | 0.108468 (rank : 15) | θ value | 0.00665767 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43567 | Gene names | RNF13, RZF | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 13. | |||||
|
RN167_MOUSE
|
||||||
| NC score | 0.106663 (rank : 16) | θ value | 0.0148317 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XF4, Q3U4S5 | Gene names | Rnf167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 167 precursor. | |||||
|
RN133_HUMAN
|
||||||
| NC score | 0.086332 (rank : 17) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WVZ7, Q8N7G7 | Gene names | RNF133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 133. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.084328 (rank : 18) | θ value | 0.00869519 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
PSL1_MOUSE
|
||||||
| NC score | 0.078480 (rank : 19) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TD49, Q3TLQ8, Q3UG60, Q80VZ6 | Gene names | Sppl2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b). | |||||
|
GOLI_HUMAN
|
||||||
| NC score | 0.073448 (rank : 20) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86XS8, Q641P9, Q6P177, Q7L2U2, Q9P0J9 | Gene names | RNF130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130). | |||||
|
GOLI_MOUSE
|
||||||
| NC score | 0.073419 (rank : 21) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VEM1, Q80VL7, Q9QZQ6 | Gene names | Rnf130, G1rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130) (G1-related zinc finger protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.071940 (rank : 22) | θ value | 0.00298849 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PSL2_HUMAN
|
||||||
| NC score | 0.071494 (rank : 23) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TCT8, Q8TAW1, Q96SZ8 | Gene names | SPPL2A, PSL2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2A (EC 3.4.23.-) (Protein SPP-like 2A) (Protein SPPL2a) (Intramembrane protease 3) (IMP3) (Presenilin-like protein 2). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.065189 (rank : 24) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.060148 (rank : 25) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.057635 (rank : 26) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.057333 (rank : 27) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
PSL2_MOUSE
|
||||||
| NC score | 0.056800 (rank : 28) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJF9, Q8R354 | Gene names | Sppl2a, Psl2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2A (EC 3.4.23.-) (SPP-like 2A protein) (Protein SPPL2a) (Intramembrane protease 3) (IMP3) (Presenilin-like protein 2). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.055312 (rank : 29) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.054323 (rank : 30) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.054028 (rank : 31) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
RP1L1_MOUSE
|
||||||
| NC score | 0.050785 (rank : 32) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
CF051_HUMAN
|
||||||
| NC score | 0.050736 (rank : 33) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969F1 | Gene names | C6orf51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf51. | |||||
|
CB010_HUMAN
|
||||||
| NC score | 0.049939 (rank : 34) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z570, Q6ZN26 | Gene names | C2orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf10. | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.048209 (rank : 35) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.047679 (rank : 36) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.046441 (rank : 37) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.044571 (rank : 38) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.042511 (rank : 39) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.041044 (rank : 40) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.040577 (rank : 41) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.039164 (rank : 42) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.038333 (rank : 43) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
SAS10_HUMAN
|
||||||
| NC score | 0.036138 (rank : 44) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.034478 (rank : 45) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.034040 (rank : 46) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.033948 (rank : 47) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.033451 (rank : 48) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
TFR1_MOUSE
|
||||||
| NC score | 0.032683 (rank : 49) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62351, Q61560 | Gene names | Tfrc, Trfr | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.032260 (rank : 50) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.032008 (rank : 51) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
POT14_HUMAN
|
||||||
| NC score | 0.029951 (rank : 52) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.029807 (rank : 53) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.029312 (rank : 54) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.027614 (rank : 55) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
RPF53_MOUSE
|
||||||
| NC score | 0.027495 (rank : 56) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K202, Q62034, Q8BQG9 | Gene names | Praf1, Paf53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase I-associated factor 53 kDa subunit (EC 2.7.7.6) (RNA polymerase I-associated factor 1). | |||||
|
FA29A_HUMAN
|
||||||
| NC score | 0.026149 (rank : 57) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.024600 (rank : 58) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.023238 (rank : 59) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
TFR1_HUMAN
|
||||||
| NC score | 0.023017 (rank : 60) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02786, Q59G55, Q9UCN0, Q9UCU5, Q9UDF9, Q9UK21 | Gene names | TFRC | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen) (T9) (p90). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.022605 (rank : 61) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.021824 (rank : 62) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
ZHX2_HUMAN
|
||||||
| NC score | 0.021731 (rank : 63) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6X8 | Gene names | ZHX2, AFR1, KIAA0854, RAF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.019737 (rank : 64) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
RAE1_HUMAN
|
||||||
| NC score | 0.019172 (rank : 65) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24386, O43732 | Gene names | CHM, REP1, TCD | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein) (TCD protein). | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.019146 (rank : 66) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.018957 (rank : 67) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
SGOL2_MOUSE
|
||||||
| NC score | 0.018590 (rank : 68) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TSY8, Q811I4, Q8C9C4, Q8CGJ4, Q9CS55 | Gene names | Sgol2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2. | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.018352 (rank : 69) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.017037 (rank : 70) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.016724 (rank : 71) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
GP158_MOUSE
|
||||||
| NC score | 0.015715 (rank : 72) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.015675 (rank : 73) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
GCDH_HUMAN
|
||||||
| NC score | 0.014879 (rank : 74) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92947, O14719 | Gene names | GCDH | |||
|
Domain Architecture |
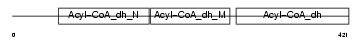
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.014309 (rank : 75) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
PPHLN_MOUSE
|
||||||
| NC score | 0.013525 (rank : 76) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2H1, Q8BUZ6 | Gene names | Pphln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.012796 (rank : 77) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.012761 (rank : 78) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.011589 (rank : 79) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.011328 (rank : 80) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SMBT1_HUMAN
|
||||||
| NC score | 0.010652 (rank : 81) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHJ3, Q96C73, Q9Y4Q9 | Gene names | SFMBT1, RU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1 (Renal ubiquitous protein 1). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.010023 (rank : 82) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.009668 (rank : 83) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.009460 (rank : 84) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.009306 (rank : 85) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.008503 (rank : 86) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.008397 (rank : 87) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.008304 (rank : 88) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MYPC2_MOUSE
|
||||||
| NC score | 0.004580 (rank : 89) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XKE0, Q8C109, Q8K2V0 | Gene names | Mybpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
EMR3_HUMAN
|
||||||
| NC score | 0.003895 (rank : 90) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY15 | Gene names | EMR3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 3 precursor (EGF-like module-containing mucin-like receptor EMR3). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.003755 (rank : 91) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
LRFN5_MOUSE
|
||||||
| NC score | 0.001167 (rank : 92) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA0, Q8BJH4, Q8BZL0 | Gene names | Lrfn5 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||