Please be patient as the page loads
|
MA1C1_HUMAN
|
||||||
| SwissProt Accessions | Q9NR34, Q9Y545 | Gene names | MAN1C1, MAN1A3, MAN1C | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IC) (Alpha-1,2-mannosidase IC) (Mannosidase alpha class 1C member 1) (HMIC). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MA1A1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990346 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33908, Q9NU44, Q9UJI3 | Gene names | MAN1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1A1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989144 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P45700 | Gene names | Man1a1, Man1a | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1C1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR34, Q9Y545 | Gene names | MAN1C1, MAN1A3, MAN1C | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IC) (Alpha-1,2-mannosidase IC) (Mannosidase alpha class 1C member 1) (HMIC). | |||||
|
MA1A2_HUMAN
|
||||||
| θ value | 1.04765e-165 (rank : 4) | NC score | 0.988076 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60476, Q9H510 | Gene names | MAN1A2, MAN1B | |||
|
Domain Architecture |
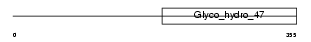
|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1A2_MOUSE
|
||||||
| θ value | 1.19867e-161 (rank : 5) | NC score | 0.988042 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39098, Q5SUY6, Q5SUY7, Q60599, Q9CUY9 | Gene names | Man1a2, Man1b | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1B1_HUMAN
|
||||||
| θ value | 1.40038e-85 (rank : 6) | NC score | 0.934773 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
EDEM2_HUMAN
|
||||||
| θ value | 6.58091e-43 (rank : 7) | NC score | 0.881438 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BV94, Q6IA89, Q6UWZ4, Q9H4U0, Q9H886, Q9NTL9, Q9NVE6 | Gene names | EDEM2, C20orf31, C20orf49 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 2 precursor. | |||||
|
EDEM1_HUMAN
|
||||||
| θ value | 2.19075e-38 (rank : 8) | NC score | 0.842247 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM1_MOUSE
|
||||||
| θ value | 6.37413e-38 (rank : 9) | NC score | 0.841424 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925U4 | Gene names | Edem1, Edem | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM3_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 10) | NC score | 0.829305 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
ZN197_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.002367 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14709 | Gene names | ZNF197, ZNF166 | |||
|
Domain Architecture |
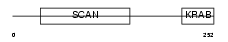
|
|||||
| Description | Zinc finger protein 197 (ZnF20). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.013395 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.002824 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ZFP28_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.001207 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |
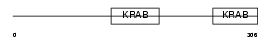
|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.012176 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
COG8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.018238 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.009761 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.010440 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | -0.000854 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
NCOAT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.008652 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.004296 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
MA1C1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR34, Q9Y545 | Gene names | MAN1C1, MAN1A3, MAN1C | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IC) (Alpha-1,2-mannosidase IC) (Mannosidase alpha class 1C member 1) (HMIC). | |||||
|
MA1A1_HUMAN
|
||||||
| NC score | 0.990346 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33908, Q9NU44, Q9UJI3 | Gene names | MAN1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1A1_MOUSE
|
||||||
| NC score | 0.989144 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P45700 | Gene names | Man1a1, Man1a | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1A2_HUMAN
|
||||||
| NC score | 0.988076 (rank : 4) | θ value | 1.04765e-165 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60476, Q9H510 | Gene names | MAN1A2, MAN1B | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1A2_MOUSE
|
||||||
| NC score | 0.988042 (rank : 5) | θ value | 1.19867e-161 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39098, Q5SUY6, Q5SUY7, Q60599, Q9CUY9 | Gene names | Man1a2, Man1b | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1B1_HUMAN
|
||||||
| NC score | 0.934773 (rank : 6) | θ value | 1.40038e-85 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
EDEM2_HUMAN
|
||||||
| NC score | 0.881438 (rank : 7) | θ value | 6.58091e-43 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BV94, Q6IA89, Q6UWZ4, Q9H4U0, Q9H886, Q9NTL9, Q9NVE6 | Gene names | EDEM2, C20orf31, C20orf49 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 2 precursor. | |||||
|
EDEM1_HUMAN
|
||||||
| NC score | 0.842247 (rank : 8) | θ value | 2.19075e-38 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM1_MOUSE
|
||||||
| NC score | 0.841424 (rank : 9) | θ value | 6.37413e-38 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925U4 | Gene names | Edem1, Edem | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM3_HUMAN
|
||||||
| NC score | 0.829305 (rank : 10) | θ value | 3.86705e-35 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
COG8_MOUSE
|
||||||
| NC score | 0.018238 (rank : 11) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.013395 (rank : 12) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.012176 (rank : 13) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.010440 (rank : 14) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.009761 (rank : 15) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NCOAT_HUMAN
|
||||||
| NC score | 0.008652 (rank : 16) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.004296 (rank : 17) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.002824 (rank : 18) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ZN197_HUMAN
|
||||||
| NC score | 0.002367 (rank : 19) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14709 | Gene names | ZNF197, ZNF166 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 197 (ZnF20). | |||||
|
ZFP28_HUMAN
|
||||||
| NC score | 0.001207 (rank : 20) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | -0.000854 (rank : 21) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||