Please be patient as the page loads
|
OST3B_HUMAN
|
||||||
| SwissProt Accessions | Q9Y662 | Gene names | HS3ST3B1, 3OST3B1, HS3ST3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (h3-OST-3B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OST3B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y662 | Gene names | HS3ST3B1, 3OST3B1, HS3ST3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (h3-OST-3B). | |||||
|
OST3B_MOUSE
|
||||||
| θ value | 7.22167e-183 (rank : 2) | NC score | 0.997337 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QZS6 | Gene names | Hs3st3b1, 3ost3b1, Hs3st3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (m3-OST-3B). | |||||
|
OST3A_HUMAN
|
||||||
| θ value | 1.46868e-151 (rank : 3) | NC score | 0.997791 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y663 | Gene names | HS3ST3A1, 3OST3A1, HS3ST3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3A1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1) (Heparan sulfate 3-O-sulfotransferase 3A1) (h3-OST-3A). | |||||
|
OST3A_MOUSE
|
||||||
| θ value | 1.09172e-138 (rank : 4) | NC score | 0.995922 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKN6 | Gene names | Hs3st3a1, 3ost3a1, Hs3st3a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3A1 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1) (Heparan sulfate 3-O-sulfotransferase 3A1). | |||||
|
OST4_HUMAN
|
||||||
| θ value | 1.87085e-122 (rank : 5) | NC score | 0.987161 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y661, Q5QI42, Q8NDC2 | Gene names | HS3ST4, 3OST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 4 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4) (Heparan sulfate 3-O-sulfotransferase 4) (h3-OST-4). | |||||
|
OST6_HUMAN
|
||||||
| θ value | 1.93603e-119 (rank : 6) | NC score | 0.994378 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96QI5, Q96RX7 | Gene names | HS3ST6, HS3ST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (h3-OST-6). | |||||
|
OST6_MOUSE
|
||||||
| θ value | 2.79562e-118 (rank : 7) | NC score | 0.994445 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5GFD5 | Gene names | Hs3st6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (3-OST-6). | |||||
|
OST2_HUMAN
|
||||||
| θ value | 1.81207e-117 (rank : 8) | NC score | 0.992903 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y278 | Gene names | HS3ST2, 3OST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 2 (EC 2.8.2.29) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 2) (Heparan sulfate 3-O-sulfotransferase 2) (h3-OST-2). | |||||
|
OST2_MOUSE
|
||||||
| θ value | 4.03687e-117 (rank : 9) | NC score | 0.992196 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q673U1, Q8BLP1, Q8C055 | Gene names | Hs3st2, 3ost2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 2 (EC 2.8.2.29) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 2) (Heparan sulfate 3-O-sulfotransferase 2). | |||||
|
OST5_HUMAN
|
||||||
| θ value | 1.36269e-64 (rank : 10) | NC score | 0.966870 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZT8, Q52LI2, Q8N285 | Gene names | HS3ST5, 3OST5, HS3OST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 5 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 5) (Heparan sulfate 3-O-sulfotransferase 5) (h3-OST-5). | |||||
|
OST5_MOUSE
|
||||||
| θ value | 1.36269e-64 (rank : 11) | NC score | 0.966835 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BSL4 | Gene names | Hs3st5, Hs3ost5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 5 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 5) (Heparan sulfate 3-O-sulfotransferase 5). | |||||
|
OST1_HUMAN
|
||||||
| θ value | 6.55044e-59 (rank : 12) | NC score | 0.966944 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14792, Q6PEY8 | Gene names | HS3ST1, 3OST, 3OST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 1 precursor (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 1) (Heparan sulfate 3-O-sulfotransferase 1) (h3-OST-1). | |||||
|
OST1_MOUSE
|
||||||
| θ value | 4.24587e-58 (rank : 13) | NC score | 0.966870 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35310 | Gene names | Hs3st1, 3ost, 3ost1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 1 precursor (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 1) (Heparan sulfate 3-O-sulfotransferase 1). | |||||
|
NDST3_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 14) | NC score | 0.759709 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95803, Q4W5C1, Q4W5D0, Q6UWC5, Q9UP21 | Gene names | NDST3, HSST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 3 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 3) (NDST- 3) (hNDST-3) (N-heparan sulfate sulfotransferase 3) (N-HSST 3) [Includes: Heparan sulfate N-deacetylase 3 (EC 3.-.-.-); Heparan sulfate N-sulfotransferase 3 (EC 2.8.2.-)]. | |||||
|
NDST4_MOUSE
|
||||||
| θ value | 1.2049e-28 (rank : 15) | NC score | 0.755551 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQW8 | Gene names | Ndst4, Hsst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
NDST3_MOUSE
|
||||||
| θ value | 1.57365e-28 (rank : 16) | NC score | 0.754885 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQH7, Q6AXE0, Q9D557 | Gene names | Ndst3, Hsst3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 3 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 3) (NDST- 3) (N-heparan sulfate sulfotransferase 3) (N-HSST 3) [Includes: Heparan sulfate N-deacetylase 3 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 3 (EC 2.8.2.-)]. | |||||
|
NDST4_HUMAN
|
||||||
| θ value | 3.50572e-28 (rank : 17) | NC score | 0.757515 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H3R1 | Gene names | NDST4, HSST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
NDST2_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 18) | NC score | 0.757022 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52849, Q2TB32, Q59H89 | Gene names | NDST2, HSST2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
NDST2_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 19) | NC score | 0.754074 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52850, Q3UDF4, Q549P5 | Gene names | Ndst2, Hsst2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) (Mndns) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N-sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
NDST1_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 20) | NC score | 0.743671 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52848, Q96E57 | Gene names | NDST1, HSST, HSST1 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 1) (NDST- 1) ([Heparan sulfate]-glucosamine N-sulfotransferase 1) (HSNST 1) (N- heparan sulfate sulfotransferase 1) (N-HSST 1) [Includes: Heparan sulfate N-deacetylase 1 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 1 (EC 2.8.2.-)]. | |||||
|
NDST1_MOUSE
|
||||||
| θ value | 1.92365e-26 (rank : 21) | NC score | 0.743084 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UHN9, O70353, Q3TBX3, Q3TDS3, Q8BZE5, Q9R206 | Gene names | Ndst1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 1) (NDST- 1) ([Heparan sulfate]-glucosamine N-sulfotransferase 1) (HSNST 1) (N- heparan sulfate sulfotransferase 1) (N-HSST 1) [Includes: Heparan sulfate N-deacetylase 1 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 1 (EC 2.8.2.-)]. | |||||
|
ST4S6_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 22) | NC score | 0.489089 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XQ5, Q80TW4, Q8BLQ5, Q9D2N6 | Gene names | Galnac4s6st, Brag, Kiaa0598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase (EC 2.8.2.33) (GalNAc4S-6ST) (B-cell RAG-associated gene protein). | |||||
|
ST4S6_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 23) | NC score | 0.470301 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7LFX5, O60338, O60474, Q86VM4 | Gene names | GALNAC4S6ST, BRAG, KIAA0598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase (EC 2.8.2.33) (GalNAc4S-6ST) (B-cell RAG-associated gene protein) (hBRAG). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 24) | NC score | 0.019135 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CHST7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.048742 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NS84, O75667 | Gene names | CHST7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 7 (EC 2.8.2.17) (EC 2.8.2.-) (Chondroitin 6-sulfotransferase 2) (C6ST-2) (N-acetylglucosamine 6-O- sulfotransferase 1) (GlcNAc6ST-4) (Galactose/N-acetylglucosamine/N- acetylglucosamine 6-O-sulfotransferase 5) (GST-5). | |||||
|
ST1C2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.041882 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75897, Q53S63 | Gene names | SULT1C2 | |||
|
Domain Architecture |

|
|||||
| Description | Sulfotransferase 1C2 (EC 2.8.2.-) (SULT1C) (SULT1C#2). | |||||
|
TPST2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.044493 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88856, Q8CBA8 | Gene names | Tpst2, D5ucla3 | |||
|
Domain Architecture |
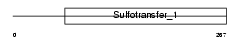
|
|||||
| Description | Protein-tyrosine sulfotransferase 2 (EC 2.8.2.20) (Tyrosylprotein sulfotransferase-2) (TPST-2). | |||||
|
ST2B1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.064273 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35400, Q8K472, Q91V03 | Gene names | Sult2b1, Sult2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase family cytosolic 2B member 1 (EC 2.8.2.2) (Sulfotransferase 2B1) (Sulfotransferase 2B) (Alcohol sulfotransferase) (Hydroxysteroid sulfotransferase 2). | |||||
|
CHST7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.046151 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EP78, Q99NB0 | Gene names | Chst7, Gst5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 7 (EC 2.8.2.17) (EC 2.8.2.-) (Chondroitin 6-sulfotransferase 2) (C6ST-2) (mC6ST-2) (N- acetylglucosamine 6-O-sulfotransferase 1) (GlcNAc6ST-4) (Galactose/N- acetylglucosamine/N-acetylglucosamine 6-O-sulfotransferase 5) (GST-5). | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.015629 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
ST2B1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.056429 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00204, O00205, O75814 | Gene names | SULT2B1, HSST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase family cytosolic 2B member 1 (EC 2.8.2.2) (Sulfotransferase 2B1) (Alcohol sulfotransferase) (Hydroxysteroid sulfotransferase 2). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.006816 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
DOK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.012552 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L591, Q8N864, Q9BQB3, Q9H666 | Gene names | DOK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.011879 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
EAF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.008766 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.002698 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.008659 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
KTNB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | -0.000158 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG40, Q8CD18, Q8R1J0, Q9CWV2 | Gene names | Katnb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p80 WD40-containing subunit B1 (Katanin p80 subunit B1) (p80 katanin). | |||||
|
LIX1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.007087 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BQ89 | Gene names | Lix1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIX1-like protein. | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.001271 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NEK9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | -0.003653 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.004739 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.005525 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.004004 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.000740 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ST1A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.014846 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52840 | Gene names | Sult1a1, St1a1, Stp, Stp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase 1A1 (EC 2.8.2.1) (Aryl sulfotransferase) (Phenol sulfotransferase) (Sulfokinase) (Phenol/aryl sulfotransferase) (mSTp1) (ST1A4). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.008123 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
SAM14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.009643 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
SGIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.009371 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SGIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.009796 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.000853 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
OST3B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y662 | Gene names | HS3ST3B1, 3OST3B1, HS3ST3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (h3-OST-3B). | |||||
|
OST3A_HUMAN
|
||||||
| NC score | 0.997791 (rank : 2) | θ value | 1.46868e-151 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y663 | Gene names | HS3ST3A1, 3OST3A1, HS3ST3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3A1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1) (Heparan sulfate 3-O-sulfotransferase 3A1) (h3-OST-3A). | |||||
|
OST3B_MOUSE
|
||||||
| NC score | 0.997337 (rank : 3) | θ value | 7.22167e-183 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QZS6 | Gene names | Hs3st3b1, 3ost3b1, Hs3st3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (m3-OST-3B). | |||||
|
OST3A_MOUSE
|
||||||
| NC score | 0.995922 (rank : 4) | θ value | 1.09172e-138 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKN6 | Gene names | Hs3st3a1, 3ost3a1, Hs3st3a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3A1 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1) (Heparan sulfate 3-O-sulfotransferase 3A1). | |||||
|
OST6_MOUSE
|
||||||
| NC score | 0.994445 (rank : 5) | θ value | 2.79562e-118 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5GFD5 | Gene names | Hs3st6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (3-OST-6). | |||||
|
OST6_HUMAN
|
||||||
| NC score | 0.994378 (rank : 6) | θ value | 1.93603e-119 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96QI5, Q96RX7 | Gene names | HS3ST6, HS3ST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (h3-OST-6). | |||||
|
OST2_HUMAN
|
||||||
| NC score | 0.992903 (rank : 7) | θ value | 1.81207e-117 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y278 | Gene names | HS3ST2, 3OST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 2 (EC 2.8.2.29) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 2) (Heparan sulfate 3-O-sulfotransferase 2) (h3-OST-2). | |||||
|
OST2_MOUSE
|
||||||
| NC score | 0.992196 (rank : 8) | θ value | 4.03687e-117 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q673U1, Q8BLP1, Q8C055 | Gene names | Hs3st2, 3ost2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 2 (EC 2.8.2.29) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 2) (Heparan sulfate 3-O-sulfotransferase 2). | |||||
|
OST4_HUMAN
|
||||||
| NC score | 0.987161 (rank : 9) | θ value | 1.87085e-122 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y661, Q5QI42, Q8NDC2 | Gene names | HS3ST4, 3OST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 4 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4) (Heparan sulfate 3-O-sulfotransferase 4) (h3-OST-4). | |||||
|
OST1_HUMAN
|
||||||
| NC score | 0.966944 (rank : 10) | θ value | 6.55044e-59 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14792, Q6PEY8 | Gene names | HS3ST1, 3OST, 3OST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 1 precursor (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 1) (Heparan sulfate 3-O-sulfotransferase 1) (h3-OST-1). | |||||
|
OST1_MOUSE
|
||||||
| NC score | 0.966870 (rank : 11) | θ value | 4.24587e-58 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35310 | Gene names | Hs3st1, 3ost, 3ost1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 1 precursor (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 1) (Heparan sulfate 3-O-sulfotransferase 1). | |||||
|
OST5_HUMAN
|
||||||
| NC score | 0.966870 (rank : 12) | θ value | 1.36269e-64 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZT8, Q52LI2, Q8N285 | Gene names | HS3ST5, 3OST5, HS3OST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 5 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 5) (Heparan sulfate 3-O-sulfotransferase 5) (h3-OST-5). | |||||
|
OST5_MOUSE
|
||||||
| NC score | 0.966835 (rank : 13) | θ value | 1.36269e-64 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BSL4 | Gene names | Hs3st5, Hs3ost5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 5 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 5) (Heparan sulfate 3-O-sulfotransferase 5). | |||||
|
NDST3_HUMAN
|
||||||
| NC score | 0.759709 (rank : 14) | θ value | 1.85889e-29 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95803, Q4W5C1, Q4W5D0, Q6UWC5, Q9UP21 | Gene names | NDST3, HSST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 3 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 3) (NDST- 3) (hNDST-3) (N-heparan sulfate sulfotransferase 3) (N-HSST 3) [Includes: Heparan sulfate N-deacetylase 3 (EC 3.-.-.-); Heparan sulfate N-sulfotransferase 3 (EC 2.8.2.-)]. | |||||
|
NDST4_HUMAN
|
||||||
| NC score | 0.757515 (rank : 15) | θ value | 3.50572e-28 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H3R1 | Gene names | NDST4, HSST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
NDST2_HUMAN
|
||||||
| NC score | 0.757022 (rank : 16) | θ value | 5.97985e-28 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52849, Q2TB32, Q59H89 | Gene names | NDST2, HSST2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
NDST4_MOUSE
|
||||||
| NC score | 0.755551 (rank : 17) | θ value | 1.2049e-28 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQW8 | Gene names | Ndst4, Hsst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
NDST3_MOUSE
|
||||||
| NC score | 0.754885 (rank : 18) | θ value | 1.57365e-28 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQH7, Q6AXE0, Q9D557 | Gene names | Ndst3, Hsst3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 3 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 3) (NDST- 3) (N-heparan sulfate sulfotransferase 3) (N-HSST 3) [Includes: Heparan sulfate N-deacetylase 3 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 3 (EC 2.8.2.-)]. | |||||
|
NDST2_MOUSE
|
||||||
| NC score | 0.754074 (rank : 19) | θ value | 7.80994e-28 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52850, Q3UDF4, Q549P5 | Gene names | Ndst2, Hsst2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) (Mndns) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N-sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
NDST1_HUMAN
|
||||||
| NC score | 0.743671 (rank : 20) | θ value | 5.06226e-27 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52848, Q96E57 | Gene names | NDST1, HSST, HSST1 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 1) (NDST- 1) ([Heparan sulfate]-glucosamine N-sulfotransferase 1) (HSNST 1) (N- heparan sulfate sulfotransferase 1) (N-HSST 1) [Includes: Heparan sulfate N-deacetylase 1 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 1 (EC 2.8.2.-)]. | |||||
|
NDST1_MOUSE
|
||||||
| NC score | 0.743084 (rank : 21) | θ value | 1.92365e-26 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UHN9, O70353, Q3TBX3, Q3TDS3, Q8BZE5, Q9R206 | Gene names | Ndst1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 1) (NDST- 1) ([Heparan sulfate]-glucosamine N-sulfotransferase 1) (HSNST 1) (N- heparan sulfate sulfotransferase 1) (N-HSST 1) [Includes: Heparan sulfate N-deacetylase 1 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 1 (EC 2.8.2.-)]. | |||||
|
ST4S6_MOUSE
|
||||||
| NC score | 0.489089 (rank : 22) | θ value | 7.59969e-07 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XQ5, Q80TW4, Q8BLQ5, Q9D2N6 | Gene names | Galnac4s6st, Brag, Kiaa0598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase (EC 2.8.2.33) (GalNAc4S-6ST) (B-cell RAG-associated gene protein). | |||||
|
ST4S6_HUMAN
|
||||||
| NC score | 0.470301 (rank : 23) | θ value | 1.69304e-06 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7LFX5, O60338, O60474, Q86VM4 | Gene names | GALNAC4S6ST, BRAG, KIAA0598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase (EC 2.8.2.33) (GalNAc4S-6ST) (B-cell RAG-associated gene protein) (hBRAG). | |||||
|
ST2B1_MOUSE
|
||||||
| NC score | 0.064273 (rank : 24) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35400, Q8K472, Q91V03 | Gene names | Sult2b1, Sult2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase family cytosolic 2B member 1 (EC 2.8.2.2) (Sulfotransferase 2B1) (Sulfotransferase 2B) (Alcohol sulfotransferase) (Hydroxysteroid sulfotransferase 2). | |||||
|
ST2B1_HUMAN
|
||||||
| NC score | 0.056429 (rank : 25) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00204, O00205, O75814 | Gene names | SULT2B1, HSST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase family cytosolic 2B member 1 (EC 2.8.2.2) (Sulfotransferase 2B1) (Alcohol sulfotransferase) (Hydroxysteroid sulfotransferase 2). | |||||
|
CHST7_HUMAN
|
||||||
| NC score | 0.048742 (rank : 26) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NS84, O75667 | Gene names | CHST7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 7 (EC 2.8.2.17) (EC 2.8.2.-) (Chondroitin 6-sulfotransferase 2) (C6ST-2) (N-acetylglucosamine 6-O- sulfotransferase 1) (GlcNAc6ST-4) (Galactose/N-acetylglucosamine/N- acetylglucosamine 6-O-sulfotransferase 5) (GST-5). | |||||
|
CHST7_MOUSE
|
||||||
| NC score | 0.046151 (rank : 27) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EP78, Q99NB0 | Gene names | Chst7, Gst5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 7 (EC 2.8.2.17) (EC 2.8.2.-) (Chondroitin 6-sulfotransferase 2) (C6ST-2) (mC6ST-2) (N- acetylglucosamine 6-O-sulfotransferase 1) (GlcNAc6ST-4) (Galactose/N- acetylglucosamine/N-acetylglucosamine 6-O-sulfotransferase 5) (GST-5). | |||||
|
TPST2_MOUSE
|
||||||
| NC score | 0.044493 (rank : 28) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88856, Q8CBA8 | Gene names | Tpst2, D5ucla3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-tyrosine sulfotransferase 2 (EC 2.8.2.20) (Tyrosylprotein sulfotransferase-2) (TPST-2). | |||||
|
ST1C2_HUMAN
|
||||||
| NC score | 0.041882 (rank : 29) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75897, Q53S63 | Gene names | SULT1C2 | |||
|
Domain Architecture |

|
|||||
| Description | Sulfotransferase 1C2 (EC 2.8.2.-) (SULT1C) (SULT1C#2). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.019135 (rank : 30) | θ value | 0.0193708 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.015629 (rank : 31) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
ST1A1_MOUSE
|
||||||
| NC score | 0.014846 (rank : 32) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52840 | Gene names | Sult1a1, St1a1, Stp, Stp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase 1A1 (EC 2.8.2.1) (Aryl sulfotransferase) (Phenol sulfotransferase) (Sulfokinase) (Phenol/aryl sulfotransferase) (mSTp1) (ST1A4). | |||||
|
DOK3_HUMAN
|
||||||
| NC score | 0.012552 (rank : 33) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L591, Q8N864, Q9BQB3, Q9H666 | Gene names | DOK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.011879 (rank : 34) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SGIP1_MOUSE
|
||||||
| NC score | 0.009796 (rank : 35) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SAM14_HUMAN
|
||||||
| NC score | 0.009643 (rank : 36) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
SGIP1_HUMAN
|
||||||
| NC score | 0.009371 (rank : 37) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
EAF1_HUMAN
|
||||||
| NC score | 0.008766 (rank : 38) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.008659 (rank : 39) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.008123 (rank : 40) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
LIX1L_MOUSE
|
||||||
| NC score | 0.007087 (rank : 41) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BQ89 | Gene names | Lix1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIX1-like protein. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.006816 (rank : 42) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.005525 (rank : 43) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.004739 (rank : 44) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.004004 (rank : 45) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.002698 (rank : 46) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.001271 (rank : 47) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.000853 (rank : 48) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.000740 (rank : 49) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
KTNB1_MOUSE
|
||||||
| NC score | -0.000158 (rank : 50) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG40, Q8CD18, Q8R1J0, Q9CWV2 | Gene names | Katnb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p80 WD40-containing subunit B1 (Katanin p80 subunit B1) (p80 katanin). | |||||
|
NEK9_HUMAN
|
||||||
| NC score | -0.003653 (rank : 51) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||