Please be patient as the page loads
|
SMG7_HUMAN
|
||||||
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMG7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984958 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
EST1A_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 3) | NC score | 0.532098 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 4) | NC score | 0.521121 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
SMG5_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 5) | NC score | 0.467036 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
SMG5_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 6) | NC score | 0.488559 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 7) | NC score | 0.055087 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.096832 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.073533 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.052149 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.081292 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.079212 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.053173 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.096616 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.077764 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.024146 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.079445 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.046108 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.041631 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
SCAM2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.041116 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15127, Q9BQE8 | Gene names | SCAMP2 | |||
|
Domain Architecture |

|
|||||
| Description | Secretory carrier-associated membrane protein 2 (Secretory carrier membrane protein 2). | |||||
|
CDC16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.092561 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CDC16_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.091981 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |
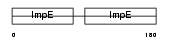
|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
DAZ2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.062977 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |
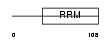
|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.012541 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.024790 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.047791 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.044695 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.048134 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.089954 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
DAZ3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.059737 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |
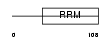
|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.012833 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.056790 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NEK4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.006695 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51957 | Gene names | NEK4, STK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2) (Serine/threonine-protein kinase NRK2). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.062058 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.029724 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.047732 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LARP4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.051947 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.032821 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.056042 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.046234 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.067345 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.041753 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
DAZ1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.053571 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |
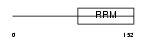
|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
DAZ4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.053608 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.022574 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.046497 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.031356 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.041520 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
PORIM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.065052 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91Z22, Q3T9F6, Q3TLL2, Q3U1R6, Q8CEX4 | Gene names | Tmem123 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.062507 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.023802 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
MTMR7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.017108 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.050772 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.041677 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.041674 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.038870 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
STAM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.012724 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
ZEP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.008790 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.059073 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ASTL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.016368 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6HA09, Q8BMA1 | Gene names | Astl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.044494 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.035483 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.054682 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.011691 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.031237 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.048478 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.010089 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PAIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.044845 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
RASF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.026610 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS23, O14571, O60539, O60710, Q9HB04, Q9HB18, Q9NS22, Q9UND4, Q9UND5 | Gene names | RASSF1, RDA32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 1. | |||||
|
SCUB3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.007821 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.010214 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.022447 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.012503 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
FOXJ3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.010095 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein J3. | |||||
|
INVS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.008586 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
KLF3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.001411 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60980 | Gene names | Klf3, Bklf | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 3 (Basic krueppel-like factor) (CACCC-box-binding protein BKLF) (TEF-2). | |||||
|
LAP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.006544 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
MNAB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.014976 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.017350 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.043206 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.025500 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
SCUB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.007499 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.046401 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
AMELX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.057204 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.043397 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CBPB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.007426 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHH6, Q5EBI3, Q9QZF0 | Gene names | Cpb2, Tafi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Carboxypeptidase R) (CPR). | |||||
|
EDA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.025385 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.019334 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
IFT88_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.019305 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
LATS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.005646 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.044021 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
ODO2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.016533 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.008273 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.013202 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.046452 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.025537 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
TBX21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.010401 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.000763 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.016433 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.035976 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.021078 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
DNMBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.009748 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.002443 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
FHL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.004607 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13642, O95212, Q13230, Q13645, Q5M7Y6, Q9NZ40, Q9UKZ8, Q9Y630 | Gene names | FHL1, SLIM1 | |||
|
Domain Architecture |
|
|||||
| Description | Four and a half LIM domains protein 1 (FHL-1) (Skeletal muscle LIM- protein 1) (SLIM 1) (SLIM). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.009446 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
K0831_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.011901 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZNE5, O94920 | Gene names | KIAA0831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
K1718_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.015202 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.005580 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.032488 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.024555 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.035816 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.029655 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
NGEF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.014481 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.018381 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.002734 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.009484 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ZN710_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | -0.000006 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
CA026_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.055489 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.052164 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SMG7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_MOUSE
|
||||||
| NC score | 0.984958 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
EST1A_HUMAN
|
||||||
| NC score | 0.532098 (rank : 3) | θ value | 4.15078e-21 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.521121 (rank : 4) | θ value | 7.0802e-21 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
SMG5_MOUSE
|
||||||
| NC score | 0.488559 (rank : 5) | θ value | 1.29331e-14 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
SMG5_HUMAN
|
||||||
| NC score | 0.467036 (rank : 6) | θ value | 1.29331e-14 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.096832 (rank : 7) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.096616 (rank : 8) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
CDC16_HUMAN
|
||||||
| NC score | 0.092561 (rank : 9) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CDC16_MOUSE
|
||||||
| NC score | 0.091981 (rank : 10) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.089954 (rank : 11) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.081292 (rank : 12) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.079445 (rank : 13) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.079212 (rank : 14) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.077764 (rank : 15) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.073533 (rank : 16) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.067345 (rank : 17) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
PORIM_MOUSE
|
||||||
| NC score | 0.065052 (rank : 18) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91Z22, Q3T9F6, Q3TLL2, Q3U1R6, Q8CEX4 | Gene names | Tmem123 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123). | |||||
|
DAZ2_HUMAN
|
||||||
| NC score | 0.062977 (rank : 19) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.062507 (rank : 20) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.062058 (rank : 21) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DAZ3_HUMAN
|
||||||
| NC score | 0.059737 (rank : 22) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.059073 (rank : 23) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
AMELX_HUMAN
|
||||||
| NC score | 0.057204 (rank : 24) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.056790 (rank : 25) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.056042 (rank : 26) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
CA026_MOUSE
|
||||||
| NC score | 0.055489 (rank : 27) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.055087 (rank : 28) | θ value | 0.00390308 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.054682 (rank : 29) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
DAZ4_HUMAN
|
||||||
| NC score | 0.053608 (rank : 30) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
DAZ1_HUMAN
|
||||||
| NC score | 0.053571 (rank : 31) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.053173 (rank : 32) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.052164 (rank : 33) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.052149 (rank : 34) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
LARP4_MOUSE
|
||||||
| NC score | 0.051947 (rank : 35) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.050772 (rank : 36) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.048478 (rank : 37) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.048134 (rank : 38) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.047791 (rank : 39) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.047732 (rank : 40) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.046497 (rank : 41) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.046452 (rank : 42) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.046401 (rank : 43) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.046234 (rank : 44) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.046108 (rank : 45) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
PAIP1_HUMAN
|
||||||
| NC score | 0.044845 (rank : 46) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.044695 (rank : 47) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.044494 (rank : 48) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.044021 (rank : 49) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.043397 (rank : 50) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.043206 (rank : 51) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.041753 (rank : 52) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.041677 (rank : 53) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.041674 (rank : 54) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.041631 (rank : 55) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.041520 (rank : 56) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SCAM2_HUMAN
|
||||||
| NC score | 0.041116 (rank : 57) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15127, Q9BQE8 | Gene names | SCAMP2 | |||
|
Domain Architecture |

|
|||||
| Description | Secretory carrier-associated membrane protein 2 (Secretory carrier membrane protein 2). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.038870 (rank : 58) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.035976 (rank : 59) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.035816 (rank : 60) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.035483 (rank : 61) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.032821 (rank : 62) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.032488 (rank : 63) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.031356 (rank : 64) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.031237 (rank : 65) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.029724 (rank : 66) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.029655 (rank : 67) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
RASF1_HUMAN
|
||||||
| NC score | 0.026610 (rank : 68) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS23, O14571, O60539, O60710, Q9HB04, Q9HB18, Q9NS22, Q9UND4, Q9UND5 | Gene names | RASSF1, RDA32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 1. | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.025537 (rank : 69) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.025500 (rank : 70) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
EDA_MOUSE
|
||||||
| NC score | 0.025385 (rank : 71) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.024790 (rank : 72) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.024555 (rank : 73) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.024146 (rank : 74) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.023802 (rank : 75) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.022574 (rank : 76) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.022447 (rank : 77) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.021078 (rank : 78) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.019334 (rank : 79) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.019305 (rank : 80) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.018381 (rank : 81) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.017350 (rank : 82) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MTMR7_MOUSE
|
||||||
| NC score | 0.017108 (rank : 83) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
ODO2_MOUSE
|
||||||
| NC score | 0.016533 (rank : 84) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.016433 (rank : 85) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
ASTL_MOUSE
|
||||||
| NC score | 0.016368 (rank : 86) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6HA09, Q8BMA1 | Gene names | Astl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
K1718_HUMAN
|
||||||
| NC score | 0.015202 (rank : 87) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MNAB_HUMAN
|
||||||
| NC score | 0.014976 (rank : 88) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
NGEF_MOUSE
|
||||||
| NC score | 0.014481 (rank : 89) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.013202 (rank : 90) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.012833 (rank : 91) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
STAM2_MOUSE
|
||||||
| NC score | 0.012724 (rank : 92) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.012541 (rank : 93) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.012503 (rank : 94) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
K0831_HUMAN
|
||||||
| NC score | 0.011901 (rank : 95) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZNE5, O94920 | Gene names | KIAA0831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.011691 (rank : 96) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TBX21_HUMAN
|
||||||
| NC score | 0.010401 (rank : 97) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.010214 (rank : 98) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
FOXJ3_MOUSE
|
||||||
| NC score | 0.010095 (rank : 99) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.010089 (rank : 100) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
DNMBP_MOUSE
|
||||||
| NC score | 0.009748 (rank : 101) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.009484 (rank : 102) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.009446 (rank : 103) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ZEP2_HUMAN
|
||||||
| NC score | 0.008790 (rank : 104) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.008586 (rank : 105) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.008273 (rank : 106) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
SCUB3_MOUSE
|
||||||
| NC score | 0.007821 (rank : 107) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SCUB3_HUMAN
|
||||||
| NC score | 0.007499 (rank : 108) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
CBPB2_MOUSE
|
||||||
| NC score | 0.007426 (rank : 109) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHH6, Q5EBI3, Q9QZF0 | Gene names | Cpb2, Tafi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Carboxypeptidase R) (CPR). | |||||
|
NEK4_HUMAN
|
||||||
| NC score | 0.006695 (rank : 110) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51957 | Gene names | NEK4, STK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2) (Serine/threonine-protein kinase NRK2). | |||||
|
LAP2_HUMAN
|
||||||
| NC score | 0.006544 (rank : 111) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
LATS1_HUMAN
|
||||||
| NC score | 0.005646 (rank : 112) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.005580 (rank : 113) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
FHL1_HUMAN
|
||||||
| NC score | 0.004607 (rank : 114) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13642, O95212, Q13230, Q13645, Q5M7Y6, Q9NZ40, Q9UKZ8, Q9Y630 | Gene names | FHL1, SLIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 1 (FHL-1) (Skeletal muscle LIM- protein 1) (SLIM 1) (SLIM). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.002734 (rank : 115) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.002443 (rank : 116) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
KLF3_MOUSE
|
||||||
| NC score | 0.001411 (rank : 117) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60980 | Gene names | Klf3, Bklf | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 3 (Basic krueppel-like factor) (CACCC-box-binding protein BKLF) (TEF-2). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.000763 (rank : 118) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN710_HUMAN
|
||||||
| NC score | -0.000006 (rank : 119) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||