Please be patient as the page loads
|
SPR1B_HUMAN
|
||||||
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPR1B_MOUSE
|
||||||
| θ value | 3.17079e-29 (rank : 1) | NC score | 0.795061 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 2.96777e-27 (rank : 3) | NC score | 0.856873 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 4) | NC score | 0.984629 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 5) | NC score | 0.616305 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 6) | NC score | 0.585563 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
SPR2D_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 7) | NC score | 0.522590 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2G_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 8) | NC score | 0.565883 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 9) | NC score | 0.561535 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPR2F_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 10) | NC score | 0.563671 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR2H_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 11) | NC score | 0.508083 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SPR2I_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 12) | NC score | 0.550234 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 13) | NC score | 0.289670 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SPR2B_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 14) | NC score | 0.471208 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SPR2E_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 15) | NC score | 0.574644 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2D_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 16) | NC score | 0.613312 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2A_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 17) | NC score | 0.700697 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35326 | Gene names | SPRR2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A (SPR-2A) (2-1). | |||||
|
SPR2B_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 18) | NC score | 0.615095 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
SPRR4_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 19) | NC score | 0.558174 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PI1, Q2M1Y7, Q5T522 | Gene names | SPRR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
SPR2F_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 20) | NC score | 0.511400 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 21) | NC score | 0.231799 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SPRR4_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 22) | NC score | 0.531274 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CGN8 | Gene names | Sprr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
SPR2K_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 23) | NC score | 0.530289 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70562 | Gene names | Sprr2k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2K. | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 24) | NC score | 0.417981 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
SPR2A_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 25) | NC score | 0.624172 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
SPR2G_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.564739 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 27) | NC score | 0.428320 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
MCSP_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 28) | NC score | 0.327524 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
LCE2B_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 29) | NC score | 0.445044 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 30) | NC score | 0.414204 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 31) | NC score | 0.416260 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 32) | NC score | 0.040016 (rank : 121) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
IGHG3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.069769 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |
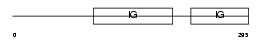
|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.229989 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.111057 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.141703 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.046783 (rank : 110) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.039706 (rank : 123) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.112359 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.065402 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.033104 (rank : 130) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.102712 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GPNMB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.052606 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P91, Q8BVV9, Q8BXL4, Q9QXA0 | Gene names | Gpnmb, Dchil, Nmb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane glycoprotein NMB precursor (Dendritic cell-associated transmembrane protein) (DC-HIL). | |||||
|
RASF7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.065500 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02833 | Gene names | RASSF7, C11orf13, HRC1 | |||
|
Domain Architecture |
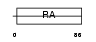
|
|||||
| Description | Ras association domain-containing protein 7 (HRAS1-related cluster protein 1). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.178154 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
CARD6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.104698 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |
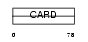
|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.036444 (rank : 127) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.034537 (rank : 128) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.037496 (rank : 126) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.072723 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.044519 (rank : 113) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.041961 (rank : 116) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
PRAX_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.209913 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
SOCS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.038352 (rank : 125) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15524, O15097, Q9NSA7 | Gene names | SOCS1, SSI1, TIP3 | |||
|
Domain Architecture |
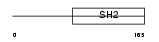
|
|||||
| Description | Suppressor of cytokine signaling 1 (SOCS-1) (JAK-binding protein) (JAB) (STAT-induced STAT inhibitor 1) (SSI-1) (Tec-interacting protein 3) (TIP-3). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.033579 (rank : 129) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.039827 (rank : 122) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
LCE1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.244328 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.156333 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.181863 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.204518 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.073943 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.085943 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.032251 (rank : 132) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
HD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.054758 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.046508 (rank : 111) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.072311 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
AMELX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.104310 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.041249 (rank : 119) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.049935 (rank : 108) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.104907 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MEF2D_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.040352 (rank : 120) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
MCSP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.231461 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.126902 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.049449 (rank : 109) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.039346 (rank : 124) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
BAI1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.013644 (rank : 143) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.059712 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.147940 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.027730 (rank : 135) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.019437 (rank : 139) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
AMELY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.069893 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
ANTR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.044443 (rank : 114) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.041600 (rank : 118) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
K1H1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.007481 (rank : 145) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61765 | Gene names | Krtha1, Hka1, Krt1-1 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha1 (Hair keratin, type I Ha1) (HKA-1). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.045057 (rank : 112) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.041916 (rank : 117) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
ODP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.025606 (rank : 136) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
SMG7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.025537 (rank : 137) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.028589 (rank : 134) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.076178 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.076879 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
AMHR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.032767 (rank : 131) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
DRD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.028947 (rank : 133) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21917 | Gene names | DRD4 | |||
|
Domain Architecture |
|
|||||
| Description | D(4) dopamine receptor (Dopamine D4 receptor) (D(2C) dopamine receptor). | |||||
|
HXA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.019016 (rank : 140) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.043673 (rank : 115) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
MOT8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.058404 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
PRR13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.146930 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SATB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.012822 (rank : 144) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01826 | Gene names | SATB1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.090373 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.017223 (rank : 141) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TRI47_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.024766 (rank : 138) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96LD4, Q96GU5, Q9BRN7 | Gene names | TRIM47, GOA, RNF100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47 (Gene overexpressed in astrocytoma protein) (RING finger protein 100). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.015264 (rank : 142) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.072043 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.060985 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.053585 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.051656 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.087766 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.067209 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.056741 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CP003_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.159602 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
FA47A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.072597 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
FA47B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.073287 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
G3PT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.052374 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64467, Q60650 | Gene names | Gapdhs, Gapd-s, Gapds | |||
|
Domain Architecture |
|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.050259 (rank : 107) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
K1802_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.064110 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KRA24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.093288 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.081327 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE3D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.163607 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYE3 | Gene names | LCE3D, LEP16, SPRL6A, SPRL6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3D (Late envelope protein 16) (Small proline-rich-like epidermal differentiation complex protein 6A) (Small proline-rich-like epidermal differentiation complex protein 6B). | |||||
|
LCE3E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.111848 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T5B0 | Gene names | LCE3E, LEP17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3E (Late envelope protein 17). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.124385 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.076969 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.057856 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.065968 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.052971 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.071110 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NOL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.054525 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |
|
|||||
| Description | Nucleolar protein 3. | |||||
|
ODFP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.089575 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |
|
|||||
| Description | Outer dense fiber protein. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.072916 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.098947 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.136703 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.052458 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.099459 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
PS1C2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.052874 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.055759 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SELV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.052863 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.066627 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SON_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.061022 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SPR2J_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.349327 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70561 | Gene names | Sprr2j | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2J. | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.076494 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.053030 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.050747 (rank : 106) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
VTNC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.051050 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
VTNC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.050975 (rank : 105) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.082571 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.090975 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.40856e-29 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.984629 (rank : 2) | θ value | 1.92365e-26 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.856873 (rank : 3) | θ value | 2.96777e-27 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.795061 (rank : 4) | θ value | 3.17079e-29 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR2A_HUMAN
|
||||||
| NC score | 0.700697 (rank : 5) | θ value | 7.1131e-05 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35326 | Gene names | SPRR2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A (SPR-2A) (2-1). | |||||
|
SPR2A_MOUSE
|
||||||
| NC score | 0.624172 (rank : 6) | θ value | 0.00665767 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.616305 (rank : 7) | θ value | 3.52202e-12 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPR2B_HUMAN
|
||||||
| NC score | 0.615095 (rank : 8) | θ value | 7.1131e-05 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
SPR2D_HUMAN
|
||||||
| NC score | 0.613312 (rank : 9) | θ value | 5.44631e-05 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.585563 (rank : 10) | θ value | 6.64225e-11 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
SPR2E_HUMAN
|
||||||
| NC score | 0.574644 (rank : 11) | θ value | 6.43352e-06 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2G_MOUSE
|
||||||
| NC score | 0.565883 (rank : 12) | θ value | 3.08544e-08 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2G_HUMAN
|
||||||
| NC score | 0.564739 (rank : 13) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
SPR2F_MOUSE
|
||||||
| NC score | 0.563671 (rank : 14) | θ value | 5.81887e-07 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.561535 (rank : 15) | θ value | 4.45536e-07 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPRR4_HUMAN
|
||||||
| NC score | 0.558174 (rank : 16) | θ value | 0.000270298 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PI1, Q2M1Y7, Q5T522 | Gene names | SPRR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
SPR2I_MOUSE
|
||||||
| NC score | 0.550234 (rank : 17) | θ value | 1.69304e-06 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
SPRR4_MOUSE
|
||||||
| NC score | 0.531274 (rank : 18) | θ value | 0.000786445 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CGN8 | Gene names | Sprr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
SPR2K_MOUSE
|
||||||
| NC score | 0.530289 (rank : 19) | θ value | 0.00228821 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70562 | Gene names | Sprr2k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2K. | |||||
|
SPR2D_MOUSE
|
||||||
| NC score | 0.522590 (rank : 20) | θ value | 1.06045e-08 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2F_HUMAN
|
||||||
| NC score | 0.511400 (rank : 21) | θ value | 0.00035302 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
SPR2H_MOUSE
|
||||||
| NC score | 0.508083 (rank : 22) | θ value | 5.81887e-07 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SPR2B_MOUSE
|
||||||
| NC score | 0.471208 (rank : 23) | θ value | 6.43352e-06 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
LCE2B_HUMAN
|
||||||
| NC score | 0.445044 (rank : 24) | θ value | 0.0148317 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.428320 (rank : 25) | θ value | 0.00869519 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.417981 (rank : 26) | θ value | 0.00665767 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.416260 (rank : 27) | θ value | 0.0148317 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.414204 (rank : 28) | θ value | 0.0148317 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
SPR2J_MOUSE
|
||||||
| NC score | 0.349327 (rank : 29) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70561 | Gene names | Sprr2j | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2J. | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.327524 (rank : 30) | θ value | 0.00869519 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.289670 (rank : 31) | θ value | 2.88788e-06 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
LCE1A_HUMAN
|
||||||
| NC score | 0.244328 (rank : 32) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.231799 (rank : 33) | θ value | 0.000461057 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MCSP_HUMAN
|
||||||
| NC score | 0.231461 (rank : 34) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.229989 (rank : 35) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.209913 (rank : 36) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.204518 (rank : 37) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.181863 (rank : 38) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.178154 (rank : 39) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
LCE3D_HUMAN
|
||||||
| NC score | 0.163607 (rank : 40) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYE3 | Gene names | LCE3D, LEP16, SPRL6A, SPRL6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3D (Late envelope protein 16) (Small proline-rich-like epidermal differentiation complex protein 6A) (Small proline-rich-like epidermal differentiation complex protein 6B). | |||||
|
CP003_HUMAN
|
||||||
| NC score | 0.159602 (rank : 41) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.156333 (rank : 42) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.147940 (rank : 43) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.146930 (rank : 44) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.141703 (rank : 45) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.136703 (rank : 46) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.126902 (rank : 47) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.124385 (rank : 48) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.112359 (rank : 49) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
LCE3E_HUMAN
|
||||||
| NC score | 0.111848 (rank : 50) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T5B0 | Gene names | LCE3E, LEP17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3E (Late envelope protein 17). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.111057 (rank : 51) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.104907 (rank : 52) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CARD6_HUMAN
|
||||||
| NC score | 0.104698 (rank : 53) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
AMELX_HUMAN
|
||||||
| NC score | 0.104310 (rank : 54) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.102712 (rank : 55) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.099459 (rank : 56) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.098947 (rank : 57) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.093288 (rank : 58) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.090975 (rank : 59) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.090373 (rank : 60) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ODFP_MOUSE
|
||||||
| NC score | 0.089575 (rank : 61) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.087766 (rank : 62) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.085943 (rank : 63) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.082571 (rank : 64) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.081327 (rank : 65) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.076969 (rank : 66) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.076879 (rank : 67) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.076494 (rank : 68) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.076178 (rank : 69) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.073943 (rank : 70) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.073287 (rank : 71) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.072916 (rank : 72) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.072723 (rank : 73) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.072597 (rank : 74) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.072311 (rank : 75) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.072043 (rank : 76) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.071110 (rank : 77) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
AMELY_HUMAN
|
||||||
| NC score | 0.069893 (rank : 78) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
IGHG3_HUMAN
|
||||||
| NC score | 0.069769 (rank : 79) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.067209 (rank : 80) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.066627 (rank : 81) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.065968 (rank : 82) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RASF7_HUMAN
|
||||||
| NC score | 0.065500 (rank : 83) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02833 | Gene names | RASSF7, C11orf13, HRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 7 (HRAS1-related cluster protein 1). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.065402 (rank : 84) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.064110 (rank : 85) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.061022 (rank : 86) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.060985 (rank : 87) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.059712 (rank : 88) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
MOT8_MOUSE
|
||||||
| NC score | 0.058404 (rank : 89) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.057856 (rank : 90) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.056741 (rank : 91) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.055759 (rank : 92) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.054758 (rank : 93) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
NOL3_MOUSE
|
||||||
| NC score | 0.054525 (rank : 94) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein 3. | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.053585 (rank : 95) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.053030 (rank : 96) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.052971 (rank : 97) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PS1C2_MOUSE
|
||||||
| NC score | 0.052874 (rank : 98) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.052863 (rank : 99) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
GPNMB_MOUSE
|
||||||
| NC score | 0.052606 (rank : 100) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P91, Q8BVV9, Q8BXL4, Q9QXA0 | Gene names | Gpnmb, Dchil, Nmb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane glycoprotein NMB precursor (Dendritic cell-associated transmembrane protein) (DC-HIL). | |||||
|
PRR8_HUMAN
|
||||||
| NC score | 0.052458 (rank : 101) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
G3PT_MOUSE
|
||||||
| NC score | 0.052374 (rank : 102) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64467, Q60650 | Gene names | Gapdhs, Gapd-s, Gapds | |||
|
Domain Architecture |

|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.051656 (rank : 103) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
VTNC_HUMAN
|
||||||
| NC score | 0.051050 (rank : 104) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.050975 (rank : 105) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.050747 (rank : 106) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.050259 (rank : 107) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.049935 (rank : 108) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.049449 (rank : 109) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.046783 (rank : 110) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.046508 (rank : 111) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.045057 (rank : 112) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.044519 (rank : 113) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
ANTR1_MOUSE
|
||||||
| NC score | 0.044443 (rank : 114) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.043673 (rank : 115) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.041961 (rank : 116) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.041916 (rank : 117) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.041600 (rank : 118) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.041249 (rank : 119) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
MEF2D_HUMAN
|
||||||
| NC score | 0.040352 (rank : 120) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.040016 (rank : 121) | θ value | 0.0193708 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.039827 (rank : 122) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.039706 (rank : 123) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.039346 (rank : 124) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
SOCS1_HUMAN
|
||||||
| NC score | 0.038352 (rank : 125) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15524, O15097, Q9NSA7 | Gene names | SOCS1, SSI1, TIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 1 (SOCS-1) (JAK-binding protein) (JAB) (STAT-induced STAT inhibitor 1) (SSI-1) (Tec-interacting protein 3) (TIP-3). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.037496 (rank : 126) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.036444 (rank : 127) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.034537 (rank : 128) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.033579 (rank : 129) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.033104 (rank : 130) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
AMHR2_HUMAN
|
||||||
| NC score | 0.032767 (rank : 131) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.032251 (rank : 132) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
DRD4_HUMAN
|
||||||
| NC score | 0.028947 (rank : 133) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21917 | Gene names | DRD4 | |||
|
Domain Architecture |

|
|||||
| Description | D(4) dopamine receptor (Dopamine D4 receptor) (D(2C) dopamine receptor). | |||||
|
SMG7_MOUSE
|
||||||
| NC score | 0.028589 (rank : 134) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.027730 (rank : 135) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ODP2_HUMAN
|
||||||
| NC score | 0.025606 (rank : 136) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
SMG7_HUMAN
|
||||||
| NC score | 0.025537 (rank : 137) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
TRI47_HUMAN
|
||||||
| NC score | 0.024766 (rank : 138) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96LD4, Q96GU5, Q9BRN7 | Gene names | TRIM47, GOA, RNF100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47 (Gene overexpressed in astrocytoma protein) (RING finger protein 100). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.019437 (rank : 139) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
HXA3_MOUSE
|
||||||
| NC score | 0.019016 (rank : 140) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.017223 (rank : 141) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.015264 (rank : 142) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
BAI1_MOUSE
|
||||||
| NC score | 0.013644 (rank : 143) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
SATB1_HUMAN
|
||||||
| NC score | 0.012822 (rank : 144) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01826 | Gene names | SATB1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
K1H1_MOUSE
|
||||||
| NC score | 0.007481 (rank : 145) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61765 | Gene names | Krtha1, Hka1, Krt1-1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha1 (Hair keratin, type I Ha1) (HKA-1). | |||||