Please be patient as the page loads
|
MEF2D_HUMAN
|
||||||
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MEF2D_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 170 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
MEF2D_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.981474 (rank : 2) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q63943, Q63944 | Gene names | Mef2d | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
MEF2A_MOUSE
|
||||||
| θ value | 1.29861e-115 (rank : 3) | NC score | 0.957492 (rank : 3) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60929 | Gene names | Mef2a | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2A. | |||||
|
MEF2C_MOUSE
|
||||||
| θ value | 4.1775e-114 (rank : 4) | NC score | 0.946014 (rank : 5) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CFN5, Q8R0H1, Q9D7L0, Q9QW20 | Gene names | Mef2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
MEF2A_HUMAN
|
||||||
| θ value | 5.45601e-114 (rank : 5) | NC score | 0.953939 (rank : 4) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
MEF2C_HUMAN
|
||||||
| θ value | 7.12574e-114 (rank : 6) | NC score | 0.943717 (rank : 6) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q06413 | Gene names | MEF2C | |||
|
Domain Architecture |
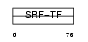
|
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
MEF2B_HUMAN
|
||||||
| θ value | 1.01529e-43 (rank : 7) | NC score | 0.881486 (rank : 7) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 5.57106e-42 (rank : 8) | NC score | 0.837552 (rank : 8) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
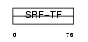
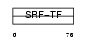
SRF_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 9) | NC score | 0.455735 (rank : 10) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P11831 | Gene names | SRF | |||
|
Domain Architecture |

|
|||||
| Description | Serum response factor (SRF). | |||||
|
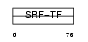
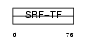
SRF_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 10) | NC score | 0.458961 (rank : 9) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JM73 | Gene names | Srf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum response factor (SRF). | |||||
|
HNF1A_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 11) | NC score | 0.079341 (rank : 16) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 12) | NC score | 0.078769 (rank : 17) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CD248_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 13) | NC score | 0.045430 (rank : 49) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.088705 (rank : 13) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.069645 (rank : 20) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.054240 (rank : 33) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.090951 (rank : 12) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.033206 (rank : 84) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.057620 (rank : 29) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
GPR98_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.031810 (rank : 91) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.053789 (rank : 34) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
EPN4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.040657 (rank : 63) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.050280 (rank : 40) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.066902 (rank : 22) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.041974 (rank : 60) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
UTX_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.038010 (rank : 71) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.050571 (rank : 39) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
EVI2B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.074243 (rank : 19) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.046993 (rank : 45) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HNF1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.057388 (rank : 30) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.057184 (rank : 31) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SPHK2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.044702 (rank : 52) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.030493 (rank : 98) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.024867 (rank : 118) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.043042 (rank : 58) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.065107 (rank : 25) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.087360 (rank : 14) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FOXP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.026977 (rank : 108) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.039358 (rank : 68) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.043327 (rank : 56) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.013490 (rank : 146) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.030571 (rank : 97) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SOX6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.029850 (rank : 101) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.023645 (rank : 121) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.026776 (rank : 110) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
CN004_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.045412 (rank : 50) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
FBN2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.009388 (rank : 157) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FOXC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.023696 (rank : 120) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |
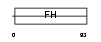
|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.026759 (rank : 111) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.093357 (rank : 11) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.043730 (rank : 53) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.065987 (rank : 24) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.035407 (rank : 76) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.013761 (rank : 143) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.033713 (rank : 82) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
JUND_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.026148 (rank : 112) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.043266 (rank : 57) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PP16A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.007330 (rank : 162) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |
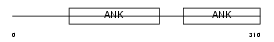
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||
|
ECM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.049036 (rank : 42) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
GAK18_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.032172 (rank : 87) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.037634 (rank : 72) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
IPMK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.032495 (rank : 86) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFU5 | Gene names | IPMK, IMPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol polyphosphate multikinase (EC 2.7.1.151) (Inositol 1,3,4,6- tetrakisphosphate 5-kinase). | |||||
|
KBTB9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.007581 (rank : 161) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80T74 | Gene names | Kbtbd9, Kiaa1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.046156 (rank : 46) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.038633 (rank : 70) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PO2F2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.013645 (rank : 144) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |
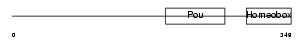
|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
RX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.013510 (rank : 145) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.039011 (rank : 69) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
WEE1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.007050 (rank : 164) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
ARRD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.031615 (rank : 92) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99KN1, Q3TDT6, Q8VEG7 | Gene names | Arrdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.055258 (rank : 32) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BOLL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.014921 (rank : 138) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N9W6, Q969U3 | Gene names | BOLL, BOULE | |||
|
Domain Architecture |
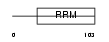
|
|||||
| Description | Protein boule-like. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.033852 (rank : 81) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.025895 (rank : 113) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
K0552_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.025314 (rank : 116) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60299 | Gene names | KIAA0552 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0552. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.063313 (rank : 26) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.082112 (rank : 15) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.052231 (rank : 36) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.059252 (rank : 28) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.020533 (rank : 128) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.015735 (rank : 136) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.013879 (rank : 142) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
DACH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.025448 (rank : 115) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.034518 (rank : 79) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.034538 (rank : 78) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.037208 (rank : 73) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
IF4G2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.031392 (rank : 93) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78344, O60877, P78404, Q53EU1, Q58EZ2, Q8NI71, Q96C16 | Gene names | EIF4G2, DAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Death-associated protein 5) (DAP-5). | |||||
|
IF4G2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.030914 (rank : 95) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62448, P97867, Q921U3 | Gene names | Eif4g2, Nat1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Novel APOBEC-1 target 1) (Translation repressor NAT1). | |||||
|
LBN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.021387 (rank : 125) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K1G2, Q8BRF3 | Gene names | Evc2, Lbn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.027344 (rank : 107) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.010797 (rank : 152) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.062219 (rank : 27) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.022107 (rank : 124) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.043586 (rank : 54) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.031906 (rank : 89) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.040352 (rank : 64) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
US6NL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.017145 (rank : 132) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.023537 (rank : 122) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.035365 (rank : 77) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.030858 (rank : 96) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.009797 (rank : 155) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.039993 (rank : 65) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.045730 (rank : 48) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.045393 (rank : 51) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.045968 (rank : 47) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.036314 (rank : 75) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
ZDHC8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.009514 (rank : 156) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |
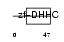
|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
ZN414_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.019883 (rank : 129) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96IQ9 | Gene names | ZNF414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.014190 (rank : 140) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.051983 (rank : 37) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
BNC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.006054 (rank : 168) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35914, O54886 | Gene names | Bnc1, Bnc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.006552 (rank : 166) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.043349 (rank : 55) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.018659 (rank : 131) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ECM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.028000 (rank : 103) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.042436 (rank : 59) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.025708 (rank : 114) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
INVS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.005265 (rank : 170) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.030217 (rank : 99) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PENK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.022159 (rank : 123) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01210 | Gene names | PENK | |||
|
Domain Architecture |

|
|||||
| Description | Proenkephalin A precursor [Contains: Synenkephalin; Met-enkephalin (Opioid growth factor) (OGF); Met-enkephalin-Arg-Gly-Leu; Leu- enkephalin; Met-enkephalin-Arg-Phe]. | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.010858 (rank : 151) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
SMR2C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.041019 (rank : 62) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35985 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform gamma precursor (Salivary protein MSG2, isoform gamma). | |||||
|
SMR2D_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.041770 (rank : 61) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35979 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform delta precursor (Salivary protein MSG2, isoform delta). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.006896 (rank : 165) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.011428 (rank : 148) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.033167 (rank : 85) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.030095 (rank : 100) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CN037_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.027427 (rank : 106) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.016356 (rank : 134) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.011295 (rank : 149) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
EPN4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.024511 (rank : 119) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |
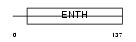
|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.017070 (rank : 133) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
HME1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.007653 (rank : 159) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09065 | Gene names | En1, En-1 | |||
|
Domain Architecture |
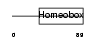
|
|||||
| Description | Homeobox protein engrailed-1 (Mo-En-1). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.033494 (rank : 83) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
LAMP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.019131 (rank : 130) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TST5, Q8C1F6 | Gene names | Lamp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosome-associated membrane glycoprotein 3 precursor (LAMP-3) (Lysosomal-associated membrane protein 3) (DC-lysosome-associated membrane glycoprotein) (DC LAMP) (CD208 antigen). | |||||
|
MB3L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.020857 (rank : 127) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.027690 (rank : 105) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.026865 (rank : 109) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
S13A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.007304 (rank : 163) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13183 | Gene names | SLC13A2, NADC1, SDCT1 | |||
|
Domain Architecture |
|
|||||
| Description | Solute carrier family 13 member 2 (Renal sodium/dicarboxylate cotransporter) (Na(+)/dicarboxylate cotransporter 1) (NaDC-1). | |||||
|
SOX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.007622 (rank : 160) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.021302 (rank : 126) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
STML1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.009864 (rank : 154) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBI4, O95675, Q6FGL8, Q8WYI7, Q9UMB9, Q9UMC0, Q9Y6H9 | Gene names | STOML1, SLP1, UNC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stomatin-like protein 1 (SLP-1) (Stomatin-related protein) (STORP) (EPB72-like 1) (UNC24 homolog). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.024892 (rank : 117) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.010408 (rank : 153) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
BTBD7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.014815 (rank : 139) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.051508 (rank : 38) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CD45_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.001801 (rank : 173) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
COR2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.002709 (rank : 172) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0P5, Q3U6H8, Q8BW42 | Gene names | Coro2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-2A. | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.036884 (rank : 74) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.047059 (rank : 44) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
FBX46_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.011112 (rank : 150) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PJ61 | Gene names | FBXO46, FBX46, FBXO34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
GAK19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.033908 (rank : 80) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.031903 (rank : 90) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.031977 (rank : 88) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GBF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.008740 (rank : 158) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.027703 (rank : 104) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
LSP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.012328 (rank : 147) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.039992 (rank : 66) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.029239 (rank : 102) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
RBM19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.005568 (rank : 169) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
ROBO3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.002840 (rank : 171) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
SC24D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.013935 (rank : 141) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.031069 (rank : 94) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.015298 (rank : 137) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | -0.000288 (rank : 175) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.015885 (rank : 135) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
VINEX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.006540 (rank : 167) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.048940 (rank : 43) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.039906 (rank : 67) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.001673 (rank : 174) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.075323 (rank : 18) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.050065 (rank : 41) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.053037 (rank : 35) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.066544 (rank : 23) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.067865 (rank : 21) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
MEF2D_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 170 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
MEF2D_MOUSE
|
||||||
| NC score | 0.981474 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q63943, Q63944 | Gene names | Mef2d | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
MEF2A_MOUSE
|
||||||
| NC score | 0.957492 (rank : 3) | θ value | 1.29861e-115 (rank : 3) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60929 | Gene names | Mef2a | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2A. | |||||
|
MEF2A_HUMAN
|
||||||
| NC score | 0.953939 (rank : 4) | θ value | 5.45601e-114 (rank : 5) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
MEF2C_MOUSE
|
||||||
| NC score | 0.946014 (rank : 5) | θ value | 4.1775e-114 (rank : 4) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CFN5, Q8R0H1, Q9D7L0, Q9QW20 | Gene names | Mef2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
MEF2C_HUMAN
|
||||||
| NC score | 0.943717 (rank : 6) | θ value | 7.12574e-114 (rank : 6) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q06413 | Gene names | MEF2C | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
MEF2B_HUMAN
|
||||||
| NC score | 0.881486 (rank : 7) | θ value | 1.01529e-43 (rank : 7) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.837552 (rank : 8) | θ value | 5.57106e-42 (rank : 8) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
SRF_MOUSE
|
||||||
| NC score | 0.458961 (rank : 9) | θ value | 7.59969e-07 (rank : 10) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JM73 | Gene names | Srf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum response factor (SRF). | |||||
|
SRF_HUMAN
|
||||||
| NC score | 0.455735 (rank : 10) | θ value | 7.59969e-07 (rank : 9) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P11831 | Gene names | SRF | |||
|
Domain Architecture |

|
|||||
| Description | Serum response factor (SRF). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.093357 (rank : 11) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.090951 (rank : 12) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.088705 (rank : 13) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.087360 (rank : 14) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.082112 (rank : 15) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
HNF1A_MOUSE
|
||||||
| NC score | 0.079341 (rank : 16) | θ value | 0.00298849 (rank : 11) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.078769 (rank : 17) | θ value | 0.00298849 (rank : 12) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.075323 (rank : 18) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
EVI2B_MOUSE
|
||||||
| NC score | 0.074243 (rank : 19) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.069645 (rank : 20) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.067865 (rank : 21) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.066902 (rank : 22) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.066544 (rank : 23) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.065987 (rank : 24) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.065107 (rank : 25) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.063313 (rank : 26) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.062219 (rank : 27) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.059252 (rank : 28) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.057620 (rank : 29) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
HNF1A_HUMAN
|
||||||
| NC score | 0.057388 (rank : 30) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.057184 (rank : 31) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.055258 (rank : 32) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.054240 (rank : 33) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.053789 (rank : 34) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.053037 (rank : 35) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.052231 (rank : 36) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.051983 (rank : 37) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.051508 (rank : 38) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.050571 (rank : 39) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.050280 (rank : 40) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.050065 (rank : 41) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
ECM1_HUMAN
|
||||||
| NC score | 0.049036 (rank : 42) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.048940 (rank : 43) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.047059 (rank : 44) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.046993 (rank : 45) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.046156 (rank : 46) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.045968 (rank : 47) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.045730 (rank : 48) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.045430 (rank : 49) | θ value | 0.00665767 (rank : 13) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.045412 (rank : 50) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.045393 (rank : 51) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SPHK2_MOUSE
|
||||||
| NC score | 0.044702 (rank : 52) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.043730 (rank : 53) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.043586 (rank : 54) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.043349 (rank : 55) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.043327 (rank : 56) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.043266 (rank : 57) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.043042 (rank : 58) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.042436 (rank : 59) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.041974 (rank : 60) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
SMR2D_MOUSE
|
||||||
| NC score | 0.041770 (rank : 61) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35979 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform delta precursor (Salivary protein MSG2, isoform delta). | |||||
|
SMR2C_MOUSE
|
||||||
| NC score | 0.041019 (rank : 62) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35985 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform gamma precursor (Salivary protein MSG2, isoform gamma). | |||||
|
EPN4_MOUSE
|
||||||
| NC score | 0.040657 (rank : 63) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.040352 (rank : 64) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.039993 (rank : 65) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.039992 (rank : 66) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.039906 (rank : 67) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.039358 (rank : 68) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.039011 (rank : 69) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.038633 (rank : 70) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
UTX_HUMAN
|
||||||
| NC score | 0.038010 (rank : 71) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.037634 (rank : 72) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.037208 (rank : 73) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.036884 (rank : 74) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.036314 (rank : 75) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.035407 (rank : 76) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.035365 (rank : 77) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.034538 (rank : 78) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.034518 (rank : 79) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK19_HUMAN
|
||||||
| NC score | 0.033908 (rank : 80) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.033852 (rank : 81) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.033713 (rank : 82) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.033494 (rank : 83) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.033206 (rank : 84) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.033167 (rank : 85) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
IPMK_HUMAN
|
||||||
| NC score | 0.032495 (rank : 86) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFU5 | Gene names | IPMK, IMPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol polyphosphate multikinase (EC 2.7.1.151) (Inositol 1,3,4,6- tetrakisphosphate 5-kinase). | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.032172 (rank : 87) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.031977 (rank : 88) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.031906 (rank : 89) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.031903 (rank : 90) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GPR98_HUMAN
|
||||||
| NC score | 0.031810 (rank : 91) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
ARRD1_MOUSE
|
||||||
| NC score | 0.031615 (rank : 92) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99KN1, Q3TDT6, Q8VEG7 | Gene names | Arrdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
IF4G2_HUMAN
|
||||||
| NC score | 0.031392 (rank : 93) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78344, O60877, P78404, Q53EU1, Q58EZ2, Q8NI71, Q96C16 | Gene names | EIF4G2, DAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Death-associated protein 5) (DAP-5). | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.031069 (rank : 94) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
IF4G2_MOUSE
|
||||||
| NC score | 0.030914 (rank : 95) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62448, P97867, Q921U3 | Gene names | Eif4g2, Nat1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Novel APOBEC-1 target 1) (Translation repressor NAT1). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.030858 (rank : 96) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.030571 (rank : 97) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.030493 (rank : 98) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.030217 (rank : 99) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.030095 (rank : 100) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
SOX6_MOUSE
|
||||||
| NC score | 0.029850 (rank : 101) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.029239 (rank : 102) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
ECM1_MOUSE
|
||||||
| NC score | 0.028000 (rank : 103) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.027703 (rank : 104) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.027690 (rank : 105) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.027427 (rank : 106) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.027344 (rank : 107) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
FOXP4_HUMAN
|
||||||
| NC score | 0.026977 (rank : 108) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.026865 (rank : 109) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.026776 (rank : 110) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.026759 (rank : 111) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.026148 (rank : 112) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.025895 (rank : 113) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.025708 (rank : 114) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
DACH1_MOUSE
|
||||||
| NC score | 0.025448 (rank : 115) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
K0552_HUMAN
|
||||||
| NC score | 0.025314 (rank : 116) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60299 | Gene names | KIAA0552 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0552. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.024892 (rank : 117) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.024867 (rank : 118) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
EPN4_HUMAN
|
||||||
| NC score | 0.024511 (rank : 119) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
FOXC1_HUMAN
|
||||||
| NC score | 0.023696 (rank : 120) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.023645 (rank : 121) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.023537 (rank : 122) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
PENK_HUMAN
|
||||||
| NC score | 0.022159 (rank : 123) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01210 | Gene names | PENK | |||
|
Domain Architecture |

|
|||||
| Description | Proenkephalin A precursor [Contains: Synenkephalin; Met-enkephalin (Opioid growth factor) (OGF); Met-enkephalin-Arg-Gly-Leu; Leu- enkephalin; Met-enkephalin-Arg-Phe]. | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.022107 (rank : 124) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
LBN_MOUSE
|
||||||
| NC score | 0.021387 (rank : 125) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K1G2, Q8BRF3 | Gene names | Evc2, Lbn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
SOX6_HUMAN
|
||||||
| NC score | 0.021302 (rank : 126) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
MB3L2_HUMAN
|
||||||
| NC score | 0.020857 (rank : 127) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.020533 (rank : 128) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
ZN414_HUMAN
|
||||||
| NC score | 0.019883 (rank : 129) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96IQ9 | Gene names | ZNF414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
LAMP3_MOUSE
|
||||||
| NC score | 0.019131 (rank : 130) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TST5, Q8C1F6 | Gene names | Lamp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosome-associated membrane glycoprotein 3 precursor (LAMP-3) (Lysosomal-associated membrane protein 3) (DC-lysosome-associated membrane glycoprotein) (DC LAMP) (CD208 antigen). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.018659 (rank : 131) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
US6NL_MOUSE
|
||||||
| NC score | 0.017145 (rank : 132) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.017070 (rank : 133) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.016356 (rank : 134) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.015885 (rank : 135) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.015735 (rank : 136) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.015298 (rank : 137) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
BOLL_HUMAN
|
||||||
| NC score | 0.014921 (rank : 138) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N9W6, Q969U3 | Gene names | BOLL, BOULE | |||
|
Domain Architecture |

|
|||||
| Description | Protein boule-like. | |||||
|
BTBD7_MOUSE
|
||||||
| NC score | 0.014815 (rank : 139) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.014190 (rank : 140) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
SC24D_HUMAN
|
||||||
| NC score | 0.013935 (rank : 141) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.013879 (rank : 142) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.013761 (rank : 143) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
PO2F2_HUMAN
|
||||||
| NC score | 0.013645 (rank : 144) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
RX_HUMAN
|
||||||
| NC score | 0.013510 (rank : 145) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.013490 (rank : 146) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
LSP1_HUMAN
|
||||||
| NC score | 0.012328 (rank : 147) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.011428 (rank : 148) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.011295 (rank : 149) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
FBX46_HUMAN
|
||||||
| NC score | 0.011112 (rank : 150) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PJ61 | Gene names | FBXO46, FBX46, FBXO34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.010858 (rank : 151) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.010797 (rank : 152) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.010408 (rank : 153) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
STML1_HUMAN
|
||||||
| NC score | 0.009864 (rank : 154) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBI4, O95675, Q6FGL8, Q8WYI7, Q9UMB9, Q9UMC0, Q9Y6H9 | Gene names | STOML1, SLP1, UNC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stomatin-like protein 1 (SLP-1) (Stomatin-related protein) (STORP) (EPB72-like 1) (UNC24 homolog). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.009797 (rank : 155) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
ZDHC8_HUMAN
|
||||||
| NC score | 0.009514 (rank : 156) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.009388 (rank : 157) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
GBF1_HUMAN
|
||||||
| NC score | 0.008740 (rank : 158) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
HME1_MOUSE
|
||||||
| NC score | 0.007653 (rank : 159) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09065 | Gene names | En1, En-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein engrailed-1 (Mo-En-1). | |||||
|
SOX2_HUMAN
|
||||||
| NC score | 0.007622 (rank : 160) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
KBTB9_MOUSE
|
||||||
| NC score | 0.007581 (rank : 161) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80T74 | Gene names | Kbtbd9, Kiaa1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
PP16A_MOUSE
|
||||||
| NC score | 0.007330 (rank : 162) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||
|
S13A2_HUMAN
|
||||||
| NC score | 0.007304 (rank : 163) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13183 | Gene names | SLC13A2, NADC1, SDCT1 | |||
|
Domain Architecture |
|
|||||
| Description | Solute carrier family 13 member 2 (Renal sodium/dicarboxylate cotransporter) (Na(+)/dicarboxylate cotransporter 1) (NaDC-1). | |||||
|
WEE1B_HUMAN
|
||||||
| NC score | 0.007050 (rank : 164) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.006896 (rank : 165) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.006552 (rank : 166) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
VINEX_MOUSE
|
||||||
| NC score | 0.006540 (rank : 167) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
BNC1_MOUSE
|
||||||
| NC score | 0.006054 (rank : 168) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35914, O54886 | Gene names | Bnc1, Bnc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
RBM19_MOUSE
|
||||||
| NC score | 0.005568 (rank : 169) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.005265 (rank : 170) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
ROBO3_MOUSE
|
||||||
| NC score | 0.002840 (rank : 171) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
COR2A_MOUSE
|
||||||
| NC score | 0.002709 (rank : 172) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0P5, Q3U6H8, Q8BW42 | Gene names | Coro2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-2A. | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.001801 (rank : 173) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.001673 (rank : 174) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
TCF8_MOUSE
|
||||||
| NC score | -0.000288 (rank : 175) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||