Please be patient as the page loads
|
SMR2D_MOUSE
|
||||||
| SwissProt Accessions | O35979 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform delta precursor (Salivary protein MSG2, isoform delta). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMR2D_MOUSE
|
||||||
| θ value | 1.00358e-83 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35979 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform delta precursor (Salivary protein MSG2, isoform delta). | |||||
|
SMR2C_MOUSE
|
||||||
| θ value | 5.69867e-79 (rank : 2) | NC score | 0.986259 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35985 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform gamma precursor (Salivary protein MSG2, isoform gamma). | |||||
|
SMR2A_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 3) | NC score | 0.773280 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09133 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform alpha precursor (Salivary protein MSG2, isoform alpha). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 4) | NC score | 0.661727 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SMR3B_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 5) | NC score | 0.546061 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 6) | NC score | 0.350078 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.095485 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ABI3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.076969 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.053075 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.096562 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
AB1IP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.075234 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.046144 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
GGN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.071149 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
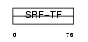
MEF2D_MOUSE
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.054200 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63943, Q63944 | Gene names | Mef2d | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
CN092_MOUSE
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.064770 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
ZBT39_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.008506 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15060 | Gene names | ZBTB39, KIAA0352 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 39. | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.047740 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.045439 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
AB1IP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.057467 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
CN092_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.058496 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
BOP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.032365 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14137, Q969Z6, Q96IS8, Q9BSA7, Q9BVM0 | Gene names | BOP1, KIAA0124 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BOP1 (Block of proliferation 1 protein). | |||||
|
LPP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.021019 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.052487 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.030713 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.026131 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TJAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.033893 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.048536 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.037327 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CPSF7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.044156 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CPSF7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.044302 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.040217 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
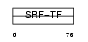
MEF2D_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.041770 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.034288 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.029358 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
ATF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.029739 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.035324 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
PCD15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.007150 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.032160 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
ARRS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.014957 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |
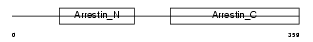
|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
BCL7C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.022728 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUZ0, O43770, Q6PD89 | Gene names | BCL7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C. | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.018186 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SMR2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.260816 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35982 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform beta precursor (Salivary protein MSG2, isoform beta). | |||||
|
SMR2E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.281777 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35961 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform epsilon precursor (Salivary protein MSG2, isoform epsilon). | |||||
|
SMR2D_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.00358e-83 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35979 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform delta precursor (Salivary protein MSG2, isoform delta). | |||||
|
SMR2C_MOUSE
|
||||||
| NC score | 0.986259 (rank : 2) | θ value | 5.69867e-79 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35985 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform gamma precursor (Salivary protein MSG2, isoform gamma). | |||||
|
SMR2A_MOUSE
|
||||||
| NC score | 0.773280 (rank : 3) | θ value | 6.85773e-16 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09133 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform alpha precursor (Salivary protein MSG2, isoform alpha). | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.661727 (rank : 4) | θ value | 2.43908e-13 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SMR3B_HUMAN
|
||||||
| NC score | 0.546061 (rank : 5) | θ value | 0.00134147 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.350078 (rank : 6) | θ value | 0.00665767 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SMR2E_MOUSE
|
||||||
| NC score | 0.281777 (rank : 7) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35961 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform epsilon precursor (Salivary protein MSG2, isoform epsilon). | |||||
|
SMR2B_MOUSE
|
||||||
| NC score | 0.260816 (rank : 8) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35982 | Gene names | Smr2, Msg2, Vcs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 2, isoform beta precursor (Salivary protein MSG2, isoform beta). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.096562 (rank : 9) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.095485 (rank : 10) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ABI3_MOUSE
|
||||||
| NC score | 0.076969 (rank : 11) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
AB1IP_MOUSE
|
||||||
| NC score | 0.075234 (rank : 12) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.071149 (rank : 13) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.064770 (rank : 14) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.058496 (rank : 15) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
AB1IP_HUMAN
|
||||||
| NC score | 0.057467 (rank : 16) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
MEF2D_MOUSE
|
||||||
| NC score | 0.054200 (rank : 17) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63943, Q63944 | Gene names | Mef2d | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.053075 (rank : 18) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.052487 (rank : 19) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.048536 (rank : 20) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.047740 (rank : 21) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.046144 (rank : 22) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.045439 (rank : 23) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
CPSF7_MOUSE
|
||||||
| NC score | 0.044302 (rank : 24) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
CPSF7_HUMAN
|
||||||
| NC score | 0.044156 (rank : 25) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
MEF2D_HUMAN
|
||||||
| NC score | 0.041770 (rank : 26) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.040217 (rank : 27) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.037327 (rank : 28) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.035324 (rank : 29) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.034288 (rank : 30) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
TJAP1_HUMAN
|
||||||
| NC score | 0.033893 (rank : 31) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
BOP1_HUMAN
|
||||||
| NC score | 0.032365 (rank : 32) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14137, Q969Z6, Q96IS8, Q9BSA7, Q9BVM0 | Gene names | BOP1, KIAA0124 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BOP1 (Block of proliferation 1 protein). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.032160 (rank : 33) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.030713 (rank : 34) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
ATF4_HUMAN
|
||||||
| NC score | 0.029739 (rank : 35) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.029358 (rank : 36) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.026131 (rank : 37) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
BCL7C_HUMAN
|
||||||
| NC score | 0.022728 (rank : 38) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUZ0, O43770, Q6PD89 | Gene names | BCL7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C. | |||||
|
LPP_MOUSE
|
||||||
| NC score | 0.021019 (rank : 39) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.018186 (rank : 40) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ARRS_HUMAN
|
||||||
| NC score | 0.014957 (rank : 41) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |

|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
ZBT39_HUMAN
|
||||||
| NC score | 0.008506 (rank : 42) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15060 | Gene names | ZBTB39, KIAA0352 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 39. | |||||
|
PCD15_MOUSE
|
||||||
| NC score | 0.007150 (rank : 43) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||