Please be patient as the page loads
|
GPNMB_MOUSE
|
||||||
| SwissProt Accessions | Q99P91, Q8BVV9, Q8BXL4, Q9QXA0 | Gene names | Gpnmb, Dchil, Nmb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane glycoprotein NMB precursor (Dendritic cell-associated transmembrane protein) (DC-HIL). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GPNMB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985487 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
GPNMB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99P91, Q8BVV9, Q8BXL4, Q9QXA0 | Gene names | Gpnmb, Dchil, Nmb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane glycoprotein NMB precursor (Dendritic cell-associated transmembrane protein) (DC-HIL). | |||||
|
PME17_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 3) | NC score | 0.753093 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P40967, Q12763, Q14448, Q14817, Q16565 | Gene names | SILV, D12S53E, PMEL17 | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Melanocyte lineage-specific antigen GP100) (Melanoma-associated ME20 antigen) (ME20M/ME20S) (ME20- M/ME20-S) (95 kDa melanocyte-specific secreted glycoprotein). | |||||
|
PME17_MOUSE
|
||||||
| θ value | 2.05049e-36 (rank : 4) | NC score | 0.777484 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 5) | NC score | 0.116106 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.051173 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.029869 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 8) | NC score | 0.048780 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.052606 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.021161 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.033489 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.023888 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MICA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.019547 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
DAF2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.016138 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |
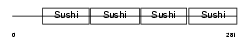
|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
GIDRP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.051335 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z5L2, O60598, Q5W0B4, Q5W0B5, Q86VT9, Q8N9H0 | Gene names | GIDRP88, C10orf28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 (Putative mitochondrial space protein 32.1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.029362 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.021951 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SON_MOUSE
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.057311 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.010128 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.013327 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.030526 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.008796 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.011738 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.012258 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.008669 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.014899 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.008988 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.028369 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.028378 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TTLL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.013306 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
AOC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.010966 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q812C9, Q80WP3 | Gene names | Aoc2 | |||
|
Domain Architecture |

|
|||||
| Description | Retina-specific copper amine oxidase precursor (EC 1.4.3.6) (RAO) (Amine oxidase [copper-containing]). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.010613 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.024925 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.028653 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PIGQ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.017552 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.001659 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.001356 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.017498 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
LEUK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.019961 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.015033 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SPR2I_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.052142 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
GPNMB_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99P91, Q8BVV9, Q8BXL4, Q9QXA0 | Gene names | Gpnmb, Dchil, Nmb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane glycoprotein NMB precursor (Dendritic cell-associated transmembrane protein) (DC-HIL). | |||||
|
GPNMB_HUMAN
|
||||||
| NC score | 0.985487 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
PME17_MOUSE
|
||||||
| NC score | 0.777484 (rank : 3) | θ value | 2.05049e-36 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
PME17_HUMAN
|
||||||
| NC score | 0.753093 (rank : 4) | θ value | 1.98146e-39 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P40967, Q12763, Q14448, Q14817, Q16565 | Gene names | SILV, D12S53E, PMEL17 | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Melanocyte lineage-specific antigen GP100) (Melanoma-associated ME20 antigen) (ME20M/ME20S) (ME20- M/ME20-S) (95 kDa melanocyte-specific secreted glycoprotein). | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.116106 (rank : 5) | θ value | 5.44631e-05 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.057311 (rank : 6) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.052606 (rank : 7) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR2I_MOUSE
|
||||||
| NC score | 0.052142 (rank : 8) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
GIDRP_HUMAN
|
||||||
| NC score | 0.051335 (rank : 9) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z5L2, O60598, Q5W0B4, Q5W0B5, Q86VT9, Q8N9H0 | Gene names | GIDRP88, C10orf28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 (Putative mitochondrial space protein 32.1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.051173 (rank : 10) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.048780 (rank : 11) | θ value | 0.47712 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.033489 (rank : 12) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.030526 (rank : 13) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.029869 (rank : 14) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.029362 (rank : 15) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.028653 (rank : 16) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.028378 (rank : 17) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.028369 (rank : 18) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.024925 (rank : 19) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.023888 (rank : 20) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.021951 (rank : 21) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.021161 (rank : 22) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.019961 (rank : 23) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
MICA2_HUMAN
|
||||||
| NC score | 0.019547 (rank : 24) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
PIGQ_HUMAN
|
||||||
| NC score | 0.017552 (rank : 25) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.017498 (rank : 26) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
DAF2_MOUSE
|
||||||
| NC score | 0.016138 (rank : 27) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.015033 (rank : 28) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.014899 (rank : 29) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.013327 (rank : 30) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.013306 (rank : 31) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.012258 (rank : 32) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.011738 (rank : 33) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
AOC2_MOUSE
|
||||||
| NC score | 0.010966 (rank : 34) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q812C9, Q80WP3 | Gene names | Aoc2 | |||
|
Domain Architecture |

|
|||||
| Description | Retina-specific copper amine oxidase precursor (EC 1.4.3.6) (RAO) (Amine oxidase [copper-containing]). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.010613 (rank : 35) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.010128 (rank : 36) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.008988 (rank : 37) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.008796 (rank : 38) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.008669 (rank : 39) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.001659 (rank : 40) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.001356 (rank : 41) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||