Please be patient as the page loads
|
SPR2E_HUMAN
|
||||||
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPR1A_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 1) | NC score | 0.470983 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 2) | NC score | 0.420426 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR2A_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 3) | NC score | 0.490003 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 4) | NC score | 0.536263 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 5) | NC score | 0.574644 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR2J_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 6) | NC score | 0.233095 (rank : 39) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70561 | Gene names | Sprr2j | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2J. | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 7) | NC score | 0.267415 (rank : 35) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 8) | NC score | 0.279272 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
SREC_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.117392 (rank : 78) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
CRDL2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.069721 (rank : 117) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6WN34, Q6WN30, Q6WN31, Q6WN32, Q7Z5J3 | Gene names | CHRDL2, BNF1, CHL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor (Chordin-related protein 2) (Breast tumor novel factor 1) (BNF-1). | |||||
|
KRA24_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.100361 (rank : 90) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
SPR2A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.884743 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35326 | Gene names | SPRR2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A (SPR-2A) (2-1). | |||||
|
TRBM_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.083465 (rank : 103) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |
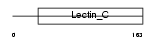
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
AMHR2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.032555 (rank : 182) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.072485 (rank : 114) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.031969 (rank : 183) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SPRR4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.445964 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PI1, Q2M1Y7, Q5T522 | Gene names | SPRR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
SPRR4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.443334 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGN8 | Gene names | Sprr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
NFX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.162173 (rank : 44) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.076469 (rank : 110) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
TRBM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.081470 (rank : 104) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |
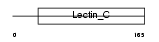
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.079841 (rank : 106) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ATS16_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.013933 (rank : 187) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TE57, Q8IVE2 | Gene names | ADAMTS16, KIAA2029 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-16 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 16) (ADAM-TS 16) (ADAM-TS16). | |||||
|
DNER_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.054854 (rank : 154) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
CC040_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.025019 (rank : 185) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q6UXB0, Q8TA84 | Gene names | C3orf40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf40 precursor. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.058536 (rank : 135) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
WIF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.062398 (rank : 130) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.064005 (rank : 126) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
ZNHI4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.028878 (rank : 184) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.048272 (rank : 181) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.017545 (rank : 186) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.098479 (rank : 94) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.053959 (rank : 158) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ALEX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.052634 (rank : 165) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
AMELX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.091844 (rank : 97) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
AMELY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.053595 (rank : 159) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
AQR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.085271 (rank : 101) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
AQR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.084891 (rank : 102) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
BMPER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.050863 (rank : 174) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
BMPER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.057003 (rank : 142) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
CARD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.072318 (rank : 116) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |
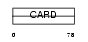
|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
CD248_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.063237 (rank : 128) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CD248_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.057427 (rank : 141) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.079423 (rank : 107) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.069551 (rank : 118) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CRDL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.051761 (rank : 170) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BU40, Q9Y3H7 | Gene names | CHRDL1, NRLN1 | |||
|
Domain Architecture |
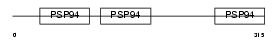
|
|||||
| Description | Chordin-like protein 1 precursor (Neuralin-1) (Ventroptin) (Neurogenesin-1). | |||||
|
CRDL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.051576 (rank : 171) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920C1, Q924K0, Q9EPZ9 | Gene names | Chrdl1, Ng1, Nrln1 | |||
|
Domain Architecture |
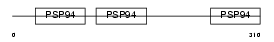
|
|||||
| Description | Chordin-like protein 1 precursor (Neuralin-1) (Ventroptin) (Neurogenesin-1). | |||||
|
CRDL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.068654 (rank : 120) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VEA6, Q925I3 | Gene names | Chrdl2, Chl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor. | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.055889 (rank : 147) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
CRUM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.050156 (rank : 179) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
DGKK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.098833 (rank : 93) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
DLK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.055028 (rank : 153) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.051295 (rank : 172) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.050403 (rank : 176) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.055865 (rank : 148) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.054814 (rank : 155) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.052082 (rank : 168) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |
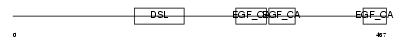
|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.052187 (rank : 167) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DNER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.055218 (rank : 150) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
EGFL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.050304 (rank : 177) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
EGFL9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050442 (rank : 175) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.057434 (rank : 140) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.063118 (rank : 129) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
G3PT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.060333 (rank : 134) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64467, Q60650 | Gene names | Gapdhs, Gapd-s, Gapds | |||
|
Domain Architecture |
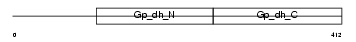
|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.053072 (rank : 162) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
IGHG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.057002 (rank : 143) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |
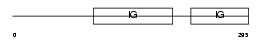
|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
JAG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.053429 (rank : 160) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.131946 (rank : 56) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR102_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.122374 (rank : 71) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR103_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.098890 (rank : 92) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR104_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.125370 (rank : 64) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR105_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.119010 (rank : 76) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.121654 (rank : 72) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR107_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.120573 (rank : 74) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR108_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.119321 (rank : 75) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR109_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.129994 (rank : 59) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KR10A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.090916 (rank : 98) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.112478 (rank : 82) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.117033 (rank : 79) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR171_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.127262 (rank : 62) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KR410_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.103287 (rank : 88) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KR412_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.132276 (rank : 54) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
KR413_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.096002 (rank : 95) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
KR414_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.123293 (rank : 68) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KR510_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.129862 (rank : 61) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KR511_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.142554 (rank : 47) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.086735 (rank : 99) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KRA13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.103336 (rank : 87) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KRA15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.068758 (rank : 119) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
KRA42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.086439 (rank : 100) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KRA44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.122434 (rank : 70) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KRA45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.144306 (rank : 46) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.139525 (rank : 50) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.110095 (rank : 84) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.111557 (rank : 83) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.129870 (rank : 60) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.124898 (rank : 66) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.133523 (rank : 53) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.100395 (rank : 89) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.125115 (rank : 65) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.131353 (rank : 57) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.123134 (rank : 69) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA92_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.132270 (rank : 55) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.130182 (rank : 58) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA94_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.124117 (rank : 67) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
KRA98_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.141509 (rank : 48) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA99_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.141155 (rank : 49) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
LCE1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.386968 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.261353 (rank : 36) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.312314 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.248968 (rank : 37) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.360920 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.369121 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.606986 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
LCE2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.613369 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.636486 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.634131 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.075282 (rank : 111) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA76 | Gene names | LCE3A, LEP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3A (Late envelope protein 13). | |||||
|
LCE3D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.183434 (rank : 40) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYE3 | Gene names | LCE3D, LEP16, SPRL6A, SPRL6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3D (Late envelope protein 16) (Small proline-rich-like epidermal differentiation complex protein 6A) (Small proline-rich-like epidermal differentiation complex protein 6B). | |||||
|
LCE3E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.127070 (rank : 63) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T5B0 | Gene names | LCE3E, LEP17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3E (Late envelope protein 17). | |||||
|
LCE4A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.099061 (rank : 91) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.602688 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.057974 (rank : 138) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.081354 (rank : 105) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MCSP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.235527 (rank : 38) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
MCSP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.481998 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.055198 (rank : 151) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.057842 (rank : 139) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25713, Q2V574 | Gene names | MT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF) (GIFB). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.062108 (rank : 132) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NAGPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.055987 (rank : 145) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NAGPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.058345 (rank : 136) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NELL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.053146 (rank : 161) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
NELL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.054257 (rank : 157) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
NELL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.055932 (rank : 146) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.050943 (rank : 173) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.050100 (rank : 180) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
ODFP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.074436 (rank : 112) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14990 | Gene names | ODF1, ODFP | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
ODFP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.137273 (rank : 52) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |
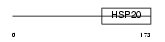
|
|||||
| Description | Outer dense fiber protein. | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.054695 (rank : 156) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
PR285_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.056560 (rank : 144) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.114075 (rank : 81) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.105016 (rank : 86) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.050276 (rank : 178) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.121120 (rank : 73) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.163786 (rank : 42) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.105915 (rank : 85) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.116523 (rank : 80) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.154007 (rank : 45) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.165816 (rank : 41) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.118246 (rank : 77) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
PS1C2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.066930 (rank : 122) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
RENT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.077782 (rank : 108) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
RENT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.077633 (rank : 109) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
SETX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.062121 (rank : 131) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.065211 (rank : 125) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.055388 (rank : 149) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.055057 (rank : 152) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.163081 (rank : 43) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SPR2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.944593 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
SPR2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.874381 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SPR2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.954109 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.891257 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.891649 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPR2F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.961370 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
SPR2F_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.919281 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR2G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.873588 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
SPR2G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.908976 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.877884 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SPR2I_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.908084 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
SPR2K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.848107 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70562 | Gene names | Sprr2k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2K. | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.377950 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.364987 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.094859 (rank : 96) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
STAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.073129 (rank : 113) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.065573 (rank : 124) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.063946 (rank : 127) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.053064 (rank : 163) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TENA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.052251 (rank : 166) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.061556 (rank : 133) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
TROAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.137922 (rank : 51) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
VWF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.058099 (rank : 137) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.052997 (rank : 164) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
WASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.051940 (rank : 169) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.066863 (rank : 123) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.067160 (rank : 121) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.072378 (rank : 115) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
SPR2E_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2F_HUMAN
|
||||||
| NC score | 0.961370 (rank : 2) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
SPR2D_HUMAN
|
||||||
| NC score | 0.954109 (rank : 3) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2B_HUMAN
|
||||||
| NC score | 0.944593 (rank : 4) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
SPR2F_MOUSE
|
||||||
| NC score | 0.919281 (rank : 5) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR2G_MOUSE
|
||||||
| NC score | 0.908976 (rank : 6) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2I_MOUSE
|
||||||
| NC score | 0.908084 (rank : 7) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.891649 (rank : 8) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPR2D_MOUSE
|
||||||
| NC score | 0.891257 (rank : 9) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2A_HUMAN
|
||||||
| NC score | 0.884743 (rank : 10) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35326 | Gene names | SPRR2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A (SPR-2A) (2-1). | |||||
|
SPR2H_MOUSE
|
||||||
| NC score | 0.877884 (rank : 11) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SPR2B_MOUSE
|
||||||
| NC score | 0.874381 (rank : 12) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SPR2G_HUMAN
|
||||||
| NC score | 0.873588 (rank : 13) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
SPR2K_MOUSE
|
||||||
| NC score | 0.848107 (rank : 14) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70562 | Gene names | Sprr2k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2K. | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.636486 (rank : 15) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.634131 (rank : 16) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE2B_HUMAN
|
||||||
| NC score | 0.613369 (rank : 17) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.606986 (rank : 18) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.602688 (rank : 19) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.574644 (rank : 20) | θ value | 6.43352e-06 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.536263 (rank : 21) | θ value | 2.88788e-06 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR2A_MOUSE
|
||||||
| NC score | 0.490003 (rank : 22) | θ value | 5.81887e-07 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.481998 (rank : 23) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.470983 (rank : 24) | θ value | 3.29651e-10 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPRR4_HUMAN
|
||||||
| NC score | 0.445964 (rank : 25) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PI1, Q2M1Y7, Q5T522 | Gene names | SPRR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
SPRR4_MOUSE
|
||||||
| NC score | 0.443334 (rank : 26) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGN8 | Gene names | Sprr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.420426 (rank : 27) | θ value | 4.45536e-07 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
LCE1A_HUMAN
|
||||||
| NC score | 0.386968 (rank : 28) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.377950 (rank : 29) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.369121 (rank : 30) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.364987 (rank : 31) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.360920 (rank : 32) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.312314 (rank : 33) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.279272 (rank : 34) | θ value | 0.00298849 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.267415 (rank : 35) | θ value | 0.00298849 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.261353 (rank : 36) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.248968 (rank : 37) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
MCSP_HUMAN
|
||||||
| NC score | 0.235527 (rank : 38) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
SPR2J_MOUSE
|
||||||
| NC score | 0.233095 (rank : 39) | θ value | 0.00134147 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70561 | Gene names | Sprr2j | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2J. | |||||
|
LCE3D_HUMAN
|
||||||
| NC score | 0.183434 (rank : 40) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYE3 | Gene names | LCE3D, LEP16, SPRL6A, SPRL6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3D (Late envelope protein 16) (Small proline-rich-like epidermal differentiation complex protein 6A) (Small proline-rich-like epidermal differentiation complex protein 6B). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.165816 (rank : 41) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.163786 (rank : 42) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.163081 (rank : 43) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
NFX1_HUMAN
|
||||||
| NC score | 0.162173 (rank : 44) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.154007 (rank : 45) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.144306 (rank : 46) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.142554 (rank : 47) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.141509 (rank : 48) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA99_HUMAN
|
||||||
| NC score | 0.141155 (rank : 49) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.139525 (rank : 50) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.137922 (rank : 51) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
ODFP_MOUSE
|
||||||
| NC score | 0.137273 (rank : 52) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.133523 (rank : 53) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KR412_HUMAN
|
||||||
| NC score | 0.132276 (rank : 54) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
KRA92_HUMAN
|
||||||
| NC score | 0.132270 (rank : 55) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.131946 (rank : 56) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.131353 (rank : 57) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.130182 (rank : 58) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.129994 (rank : 59) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.129870 (rank : 60) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.129862 (rank : 61) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.127262 (rank : 62) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
LCE3E_HUMAN
|
||||||
| NC score | 0.127070 (rank : 63) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T5B0 | Gene names | LCE3E, LEP17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3E (Late envelope protein 17). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.125370 (rank : 64) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.125115 (rank : 65) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.124898 (rank : 66) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA94_HUMAN
|
||||||
| NC score | 0.124117 (rank : 67) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
KR414_HUMAN
|
||||||
| NC score | 0.123293 (rank : 68) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.123134 (rank : 69) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA44_HUMAN
|
||||||
| NC score | 0.122434 (rank : 70) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.122374 (rank : 71) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.121654 (rank : 72) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.121120 (rank : 73) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.120573 (rank : 74) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.119321 (rank : 75) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.119010 (rank : 76) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.118246 (rank : 77) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.117392 (rank : 78) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.117033 (rank : 79) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.116523 (rank : 80) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.114075 (rank : 81) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.112478 (rank : 82) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.111557 (rank : 83) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.110095 (rank : 84) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.105915 (rank : 85) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.105016 (rank : 86) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
KRA13_HUMAN
|
||||||
| NC score | 0.103336 (rank : 87) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KR410_HUMAN
|
||||||
| NC score | 0.103287 (rank : 88) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.100395 (rank : 89) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.100361 (rank : 90) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
LCE4A_HUMAN
|
||||||
| NC score | 0.099061 (rank : 91) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.098890 (rank : 92) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.098833 (rank : 93) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.098479 (rank : 94) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
KR413_HUMAN
|
||||||
| NC score | 0.096002 (rank : 95) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.094859 (rank : 96) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
AMELX_HUMAN
|
||||||
| NC score | 0.091844 (rank : 97) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
KR10A_HUMAN
|
||||||
| NC score | 0.090916 (rank : 98) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
KRA11_HUMAN
|
||||||
| NC score | 0.086735 (rank : 99) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KRA42_HUMAN
|
||||||
| NC score | 0.086439 (rank : 100) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
AQR_HUMAN
|
||||||
| NC score | 0.085271 (rank : 101) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
AQR_MOUSE
|
||||||
| NC score | 0.084891 (rank : 102) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
TRBM_MOUSE
|
||||||
| NC score | 0.083465 (rank : 103) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
TRBM_HUMAN
|
||||||
| NC score | 0.081470 (rank : 104) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.081354 (rank : 105) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.079841 (rank : 106) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.079423 (rank : 107) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
RENT1_HUMAN
|
||||||
| NC score | 0.077782 (rank : 108) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
RENT1_MOUSE
|
||||||
| NC score | 0.077633 (rank : 109) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.076469 (rank : 110) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
LCE3A_HUMAN
|
||||||
| NC score | 0.075282 (rank : 111) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA76 | Gene names | LCE3A, LEP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3A (Late envelope protein 13). | |||||
|
ODFP_HUMAN
|
||||||
| NC score | 0.074436 (rank : 112) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14990 | Gene names | ODF1, ODFP | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.073129 (rank : 113) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.072485 (rank : 114) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.072378 (rank : 115) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CARD6_HUMAN
|
||||||
| NC score | 0.072318 (rank : 116) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
CRDL2_HUMAN
|
||||||
| NC score | 0.069721 (rank : 117) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6WN34, Q6WN30, Q6WN31, Q6WN32, Q7Z5J3 | Gene names | CHRDL2, BNF1, CHL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor (Chordin-related protein 2) (Breast tumor novel factor 1) (BNF-1). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.069551 (rank : 118) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
KRA15_HUMAN
|
||||||
| NC score | 0.068758 (rank : 119) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
CRDL2_MOUSE
|
||||||
| NC score | 0.068654 (rank : 120) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VEA6, Q925I3 | Gene names | Chrdl2, Chl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.067160 (rank : 121) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
PS1C2_MOUSE
|
||||||
| NC score | 0.066930 (rank : 122) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.066863 (rank : 123) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.065573 (rank : 124) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.065211 (rank : 125) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.064005 (rank : 126) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.063946 (rank : 127) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.063237 (rank : 128) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.063118 (rank : 129) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
WIF1_HUMAN
|
||||||
| NC score | 0.062398 (rank : 130) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.062121 (rank : 131) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.062108 (rank : 132) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.061556 (rank : 133) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
G3PT_MOUSE
|
||||||
| NC score | 0.060333 (rank : 134) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64467, Q60650 | Gene names | Gapdhs, Gapd-s, Gapds | |||
|
Domain Architecture |

|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.058536 (rank : 135) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
NAGPA_MOUSE
|
||||||
| NC score | 0.058345 (rank : 136) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.058099 (rank : 137) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.057974 (rank : 138) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MT3_HUMAN
|
||||||
| NC score | 0.057842 (rank : 139) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25713, Q2V574 | Gene names | MT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF) (GIFB). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.057434 (rank : 140) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.057427 (rank : 141) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
BMPER_MOUSE
|
||||||
| NC score | 0.057003 (rank : 142) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
IGHG3_HUMAN
|
||||||
| NC score | 0.057002 (rank : 143) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
PR285_HUMAN
|
||||||
| NC score | 0.056560 (rank : 144) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
NAGPA_HUMAN
|
||||||
| NC score | 0.055987 (rank : 145) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NELL2_MOUSE
|
||||||
| NC score | 0.055932 (rank : 146) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.055889 (rank : 147) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.055865 (rank : 148) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.055388 (rank : 149) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
DNER_MOUSE
|
||||||
| NC score | 0.055218 (rank : 150) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.055198 (rank : 151) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.055057 (rank : 152) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
DLK_HUMAN
|
||||||
| NC score | 0.055028 (rank : 153) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DNER_HUMAN
|
||||||
| NC score | 0.054854 (rank : 154) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.054814 (rank : 155) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.054695 (rank : 156) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
NELL2_HUMAN
|
||||||
| NC score | 0.054257 (rank : 157) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.053959 (rank : 158) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
AMELY_HUMAN
|
||||||
| NC score | 0.053595 (rank : 159) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.053429 (rank : 160) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
NELL1_HUMAN
|
||||||
| NC score | 0.053146 (rank : 161) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.053072 (rank : 162) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.053064 (rank : 163) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.052997 (rank : 164) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.052634 (rank : 165) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.052251 (rank : 166) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.052187 (rank : 167) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL4_HUMAN
|
||||||
| NC score | 0.052082 (rank : 168) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.051940 (rank : 169) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
CRDL1_HUMAN
|
||||||
| NC score | 0.051761 (rank : 170) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BU40, Q9Y3H7 | Gene names | CHRDL1, NRLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Chordin-like protein 1 precursor (Neuralin-1) (Ventroptin) (Neurogenesin-1). | |||||
|
CRDL1_MOUSE
|
||||||
| NC score | 0.051576 (rank : 171) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920C1, Q924K0, Q9EPZ9 | Gene names | Chrdl1, Ng1, Nrln1 | |||
|
Domain Architecture |

|
|||||
| Description | Chordin-like protein 1 precursor (Neuralin-1) (Ventroptin) (Neurogenesin-1). | |||||
|
DLK_MOUSE
|
||||||
| NC score | 0.051295 (rank : 172) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.050943 (rank : 173) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
BMPER_HUMAN
|
||||||
| NC score | 0.050863 (rank : 174) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
EGFL9_MOUSE
|
||||||
| NC score | 0.050442 (rank : 175) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.050403 (rank : 176) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
EGFL9_HUMAN
|
||||||
| NC score | 0.050304 (rank : 177) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.050276 (rank : 178) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
CRUM2_HUMAN
|
||||||
| NC score | 0.050156 (rank : 179) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.050100 (rank : 180) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.048272 (rank : 181) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
AMHR2_HUMAN
|
||||||
| NC score | 0.032555 (rank : 182) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.031969 (rank : 183) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ZNHI4_MOUSE
|
||||||
| NC score | 0.028878 (rank : 184) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
CC040_HUMAN
|
||||||
| NC score | 0.025019 (rank : 185) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q6UXB0, Q8TA84 | Gene names | C3orf40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf40 precursor. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.017545 (rank : 186) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ATS16_HUMAN
|
||||||
| NC score | 0.013933 (rank : 187) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TE57, Q8IVE2 | Gene names | ADAMTS16, KIAA2029 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-16 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 16) (ADAM-TS 16) (ADAM-TS16). | |||||