Please be patient as the page loads
|
SPR2K_MOUSE
|
||||||
| SwissProt Accessions | O70562 | Gene names | Sprr2k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2K. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPR2A_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 1) | NC score | 0.522078 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 2) | NC score | 0.434703 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPR2J_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 3) | NC score | 0.336520 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70561 | Gene names | Sprr2j | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2J. | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 4) | NC score | 0.389829 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 5) | NC score | 0.492312 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 6) | NC score | 0.530289 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR2K_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70562 | Gene names | Sprr2k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2K. | |||||
|
WFDC3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.127000 (rank : 41) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
ATS13_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.056621 (rank : 106) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
TRBM_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.085296 (rank : 60) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |
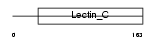
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
SPRR4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.474070 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGN8 | Gene names | Sprr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
TRBM_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.087352 (rank : 54) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |
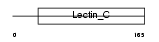
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
AMHR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.033195 (rank : 124) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
KR124_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.046254 (rank : 122) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
KRA24_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.084114 (rank : 61) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
PPAL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.092445 (rank : 51) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11117, Q9BTU7 | Gene names | ACP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal acid phosphatase precursor (EC 3.1.3.2) (LAP). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.050056 (rank : 119) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.046944 (rank : 121) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SPRR4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.433671 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PI1, Q2M1Y7, Q5T522 | Gene names | SPRR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.034776 (rank : 123) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
PPAL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.089833 (rank : 53) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24638, Q8QZT5 | Gene names | Acp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal acid phosphatase precursor (EC 3.1.3.2) (LAP). | |||||
|
SORC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.023210 (rank : 125) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.047793 (rank : 120) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.053817 (rank : 111) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.097725 (rank : 48) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
CHST7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.019085 (rank : 127) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS84, O75667 | Gene names | CHST7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 7 (EC 2.8.2.17) (EC 2.8.2.-) (Chondroitin 6-sulfotransferase 2) (C6ST-2) (N-acetylglucosamine 6-O- sulfotransferase 1) (GlcNAc6ST-4) (Galactose/N-acetylglucosamine/N- acetylglucosamine 6-O-sulfotransferase 5) (GST-5). | |||||
|
DEDD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.022787 (rank : 126) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.082706 (rank : 66) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
AMELX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.076710 (rank : 78) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
ATS13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.057326 (rank : 105) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CARD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.064503 (rank : 95) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |
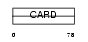
|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
CD248_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.057590 (rank : 104) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CD248_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.051348 (rank : 116) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.063201 (rank : 98) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.055540 (rank : 108) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
DGKK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.083939 (rank : 62) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
G3PT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.051614 (rank : 114) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64467, Q60650 | Gene names | Gapdhs, Gapd-s, Gapds | |||
|
Domain Architecture |
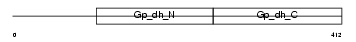
|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
IGHG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.050199 (rank : 117) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |
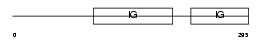
|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.062406 (rank : 99) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR102_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.076711 (rank : 77) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR103_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.054368 (rank : 110) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR104_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.083324 (rank : 65) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR105_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.068316 (rank : 88) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.076768 (rank : 76) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR107_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.078338 (rank : 73) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR108_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.081623 (rank : 69) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR109_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.067965 (rank : 90) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.060525 (rank : 103) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.076634 (rank : 79) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.061536 (rank : 100) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59990 | Gene names | KRTAP12-1, KAP12.1, KRTAP12.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-1 (Keratin-associated protein 12.1) (High sulfur keratin-associated protein 12.1). | |||||
|
KR171_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.086168 (rank : 57) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KR410_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.065895 (rank : 93) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KR412_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.083352 (rank : 64) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
KR413_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.070269 (rank : 86) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
KR414_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.082411 (rank : 67) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KR510_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.074467 (rank : 81) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KR511_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.081652 (rank : 68) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.055283 (rank : 109) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KRA13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.061097 (rank : 102) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KRA15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.064339 (rank : 97) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
KRA42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.064378 (rank : 96) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KRA44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.086583 (rank : 55) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KRA45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.083676 (rank : 63) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.080607 (rank : 70) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.061375 (rank : 101) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.067332 (rank : 91) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.071231 (rank : 84) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.068065 (rank : 89) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.069743 (rank : 87) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.070983 (rank : 85) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.073922 (rank : 83) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.077138 (rank : 75) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.065066 (rank : 94) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA92_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.075454 (rank : 80) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.077808 (rank : 74) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA94_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.078909 (rank : 72) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
KRA98_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.085710 (rank : 58) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA99_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.080342 (rank : 71) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
LCE1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.301492 (rank : 31) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.159847 (rank : 37) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.230498 (rank : 34) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.113259 (rank : 45) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.270770 (rank : 33) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.287433 (rank : 32) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.491982 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
LCE2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.488481 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.469660 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.487633 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE3D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.165423 (rank : 36) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYE3 | Gene names | LCE3D, LEP16, SPRL6A, SPRL6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3D (Late envelope protein 16) (Small proline-rich-like epidermal differentiation complex protein 6A) (Small proline-rich-like epidermal differentiation complex protein 6B). | |||||
|
LCE3E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.114933 (rank : 44) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T5B0 | Gene names | LCE3E, LEP17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3E (Late envelope protein 17). | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.462464 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.067306 (rank : 92) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MCSP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.226115 (rank : 35) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
MCSP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.410035 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
NFX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.050094 (rank : 118) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
ODFP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.056272 (rank : 107) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14990 | Gene names | ODF1, ODFP | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
ODFP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.111449 (rank : 46) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |
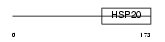
|
|||||
| Description | Outer dense fiber protein. | |||||
|
PPAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.074338 (rank : 82) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15309 | Gene names | ACPP | |||
|
Domain Architecture |

|
|||||
| Description | Prostatic acid phosphatase precursor (EC 3.1.3.2). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.092461 (rank : 50) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.085536 (rank : 59) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.097446 (rank : 49) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.134665 (rank : 40) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.086466 (rank : 56) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.091824 (rank : 52) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.126528 (rank : 42) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.153855 (rank : 38) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.099619 (rank : 47) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.051441 (rank : 115) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.142963 (rank : 39) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SPR2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.795653 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35326 | Gene names | SPRR2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A (SPR-2A) (2-1). | |||||
|
SPR2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.845587 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
SPR2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.789521 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SPR2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.838501 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.773595 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.848107 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.787987 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPR2F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.819613 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
SPR2F_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.824624 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR2G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.849375 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
SPR2G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.795824 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.757495 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SPR2I_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.805808 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.332211 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.320633 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.119810 (rank : 43) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052754 (rank : 113) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.053457 (rank : 112) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
SPR2K_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70562 | Gene names | Sprr2k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2K. | |||||
|
SPR2G_HUMAN
|
||||||
| NC score | 0.849375 (rank : 2) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
SPR2E_HUMAN
|
||||||
| NC score | 0.848107 (rank : 3) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2B_HUMAN
|
||||||
| NC score | 0.845587 (rank : 4) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
SPR2D_HUMAN
|
||||||
| NC score | 0.838501 (rank : 5) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2F_MOUSE
|
||||||
| NC score | 0.824624 (rank : 6) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR2F_HUMAN
|
||||||
| NC score | 0.819613 (rank : 7) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
SPR2I_MOUSE
|
||||||
| NC score | 0.805808 (rank : 8) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
SPR2G_MOUSE
|
||||||
| NC score | 0.795824 (rank : 9) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2A_HUMAN
|
||||||
| NC score | 0.795653 (rank : 10) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35326 | Gene names | SPRR2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A (SPR-2A) (2-1). | |||||
|
SPR2B_MOUSE
|
||||||
| NC score | 0.789521 (rank : 11) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.787987 (rank : 12) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPR2D_MOUSE
|
||||||
| NC score | 0.773595 (rank : 13) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2H_MOUSE
|
||||||
| NC score | 0.757495 (rank : 14) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.530289 (rank : 15) | θ value | 0.00228821 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR2A_MOUSE
|
||||||
| NC score | 0.522078 (rank : 16) | θ value | 4.0297e-08 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.492312 (rank : 17) | θ value | 0.000461057 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.491982 (rank : 18) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
LCE2B_HUMAN
|
||||||
| NC score | 0.488481 (rank : 19) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.487633 (rank : 20) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
SPRR4_MOUSE
|
||||||
| NC score | 0.474070 (rank : 21) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGN8 | Gene names | Sprr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.469660 (rank : 22) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.462464 (rank : 23) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.434703 (rank : 24) | θ value | 4.45536e-07 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPRR4_HUMAN
|
||||||
| NC score | 0.433671 (rank : 25) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PI1, Q2M1Y7, Q5T522 | Gene names | SPRR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.410035 (rank : 26) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.389829 (rank : 27) | θ value | 0.000158464 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR2J_MOUSE
|
||||||
| NC score | 0.336520 (rank : 28) | θ value | 9.29e-05 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70561 | Gene names | Sprr2j | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2J. | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.332211 (rank : 29) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.320633 (rank : 30) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
LCE1A_HUMAN
|
||||||
| NC score | 0.301492 (rank : 31) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.287433 (rank : 32) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.270770 (rank : 33) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.230498 (rank : 34) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
MCSP_HUMAN
|
||||||
| NC score | 0.226115 (rank : 35) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
LCE3D_HUMAN
|
||||||
| NC score | 0.165423 (rank : 36) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYE3 | Gene names | LCE3D, LEP16, SPRL6A, SPRL6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3D (Late envelope protein 16) (Small proline-rich-like epidermal differentiation complex protein 6A) (Small proline-rich-like epidermal differentiation complex protein 6B). | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.159847 (rank : 37) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.153855 (rank : 38) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.142963 (rank : 39) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.134665 (rank : 40) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
WFDC3_HUMAN
|
||||||
| NC score | 0.127000 (rank : 41) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.126528 (rank : 42) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.119810 (rank : 43) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
LCE3E_HUMAN
|
||||||
| NC score | 0.114933 (rank : 44) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T5B0 | Gene names | LCE3E, LEP17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3E (Late envelope protein 17). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.113259 (rank : 45) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
ODFP_MOUSE
|
||||||
| NC score | 0.111449 (rank : 46) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.099619 (rank : 47) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.097725 (rank : 48) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.097446 (rank : 49) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.092461 (rank : 50) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PPAL_HUMAN
|
||||||
| NC score | 0.092445 (rank : 51) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11117, Q9BTU7 | Gene names | ACP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal acid phosphatase precursor (EC 3.1.3.2) (LAP). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.091824 (rank : 52) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PPAL_MOUSE
|
||||||
| NC score | 0.089833 (rank : 53) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24638, Q8QZT5 | Gene names | Acp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal acid phosphatase precursor (EC 3.1.3.2) (LAP). | |||||
|
TRBM_HUMAN
|
||||||
| NC score | 0.087352 (rank : 54) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
KRA44_HUMAN
|
||||||
| NC score | 0.086583 (rank : 55) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.086466 (rank : 56) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.086168 (rank : 57) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.085710 (rank : 58) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.085536 (rank : 59) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
TRBM_MOUSE
|
||||||
| NC score | 0.085296 (rank : 60) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.084114 (rank : 61) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.083939 (rank : 62) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.083676 (rank : 63) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KR412_HUMAN
|
||||||
| NC score | 0.083352 (rank : 64) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.083324 (rank : 65) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.082706 (rank : 66) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
KR414_HUMAN
|
||||||
| NC score | 0.082411 (rank : 67) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.081652 (rank : 68) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.081623 (rank : 69) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.080607 (rank : 70) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA99_HUMAN
|
||||||
| NC score | 0.080342 (rank : 71) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
KRA94_HUMAN
|
||||||
| NC score | 0.078909 (rank : 72) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.078338 (rank : 73) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.077808 (rank : 74) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.077138 (rank : 75) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.076768 (rank : 76) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.076711 (rank : 77) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
AMELX_HUMAN
|
||||||
| NC score | 0.076710 (rank : 78) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.076634 (rank : 79) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KRA92_HUMAN
|
||||||
| NC score | 0.075454 (rank : 80) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.074467 (rank : 81) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
PPAP_HUMAN
|
||||||
| NC score | 0.074338 (rank : 82) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15309 | Gene names | ACPP | |||
|
Domain Architecture |

|
|||||
| Description | Prostatic acid phosphatase precursor (EC 3.1.3.2). | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.073922 (rank : 83) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.071231 (rank : 84) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.070983 (rank : 85) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KR413_HUMAN
|
||||||
| NC score | 0.070269 (rank : 86) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.069743 (rank : 87) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.068316 (rank : 88) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.068065 (rank : 89) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.067965 (rank : 90) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.067332 (rank : 91) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.067306 (rank : 92) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
KR410_HUMAN
|
||||||
| NC score | 0.065895 (rank : 93) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.065066 (rank : 94) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
CARD6_HUMAN
|
||||||
| NC score | 0.064503 (rank : 95) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
KRA42_HUMAN
|
||||||
| NC score | 0.064378 (rank : 96) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KRA15_HUMAN
|
||||||
| NC score | 0.064339 (rank : 97) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.063201 (rank : 98) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.062406 (rank : 99) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR121_HUMAN
|
||||||
| NC score | 0.061536 (rank : 100) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59990 | Gene names | KRTAP12-1, KAP12.1, KRTAP12.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-1 (Keratin-associated protein 12.1) (High sulfur keratin-associated protein 12.1). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.061375 (rank : 101) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA13_HUMAN
|
||||||
| NC score | 0.061097 (rank : 102) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.060525 (rank : 103) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.057590 (rank : 104) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.057326 (rank : 105) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
ATS13_MOUSE
|
||||||
| NC score | 0.056621 (rank : 106) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
ODFP_HUMAN
|
||||||
| NC score | 0.056272 (rank : 107) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14990 | Gene names | ODF1, ODFP | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.055540 (rank : 108) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
KRA11_HUMAN
|
||||||
| NC score | 0.055283 (rank : 109) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.054368 (rank : 110) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.053817 (rank : 111) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.053457 (rank : 112) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.052754 (rank : 113) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
G3PT_MOUSE
|
||||||
| NC score | 0.051614 (rank : 114) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64467, Q60650 | Gene names | Gapdhs, Gapd-s, Gapds | |||
|
Domain Architecture |

|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.051441 (rank : 115) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.051348 (rank : 116) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
IGHG3_HUMAN
|
||||||
| NC score | 0.050199 (rank : 117) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
NFX1_HUMAN
|
||||||
| NC score | 0.050094 (rank : 118) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.050056 (rank : 119) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.047793 (rank : 120) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.046944 (rank : 121) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
KR124_HUMAN
|
||||||
| NC score | 0.046254 (rank : 122) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.034776 (rank : 123) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
AMHR2_HUMAN
|
||||||
| NC score | 0.033195 (rank : 124) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
SORC1_MOUSE
|
||||||
| NC score | 0.023210 (rank : 125) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
DEDD_HUMAN
|
||||||
| NC score | 0.022787 (rank : 126) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
CHST7_HUMAN
|
||||||
| NC score | 0.019085 (rank : 127) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS84, O75667 | Gene names | CHST7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 7 (EC 2.8.2.17) (EC 2.8.2.-) (Chondroitin 6-sulfotransferase 2) (C6ST-2) (N-acetylglucosamine 6-O- sulfotransferase 1) (GlcNAc6ST-4) (Galactose/N-acetylglucosamine/N- acetylglucosamine 6-O-sulfotransferase 5) (GST-5). | |||||